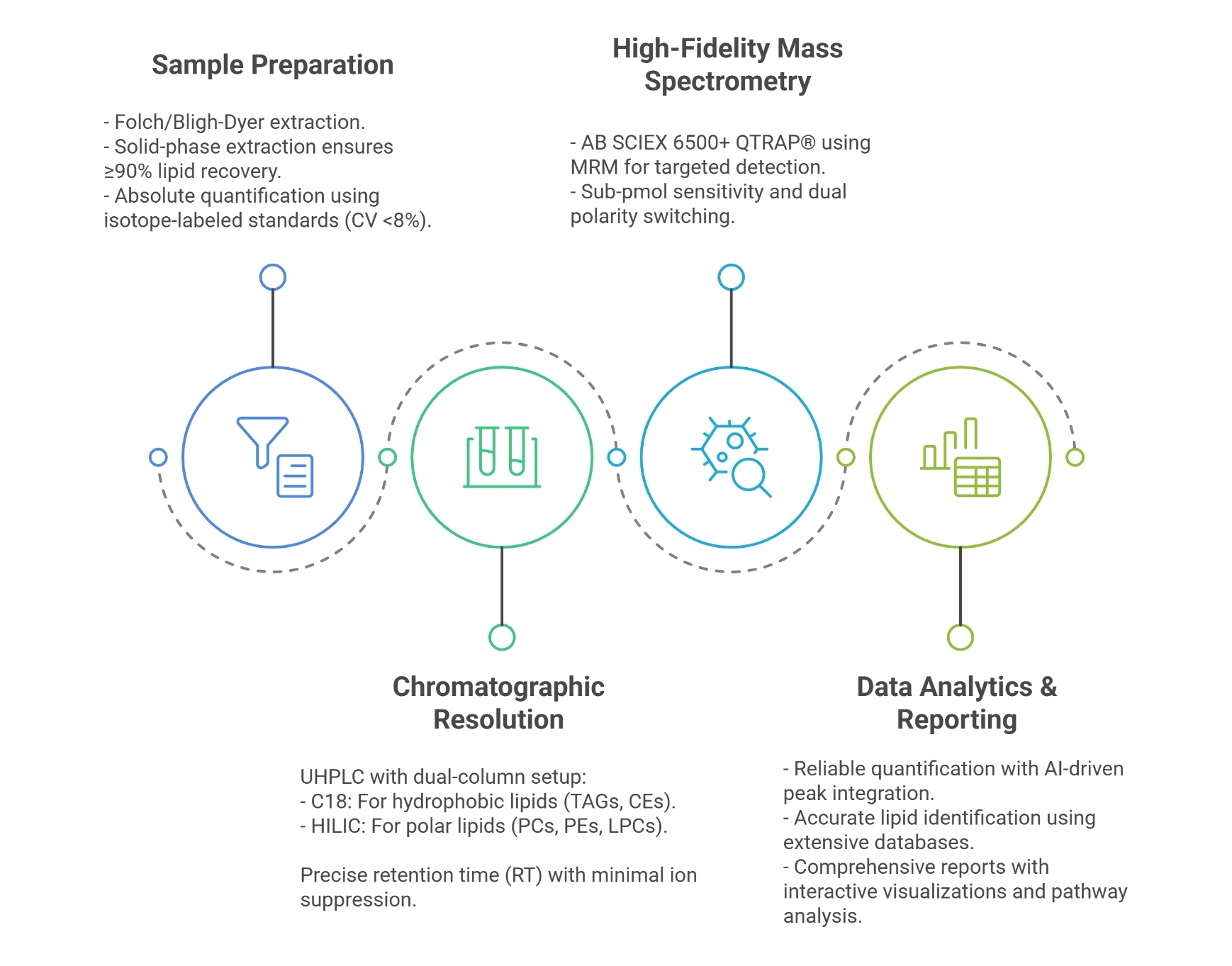
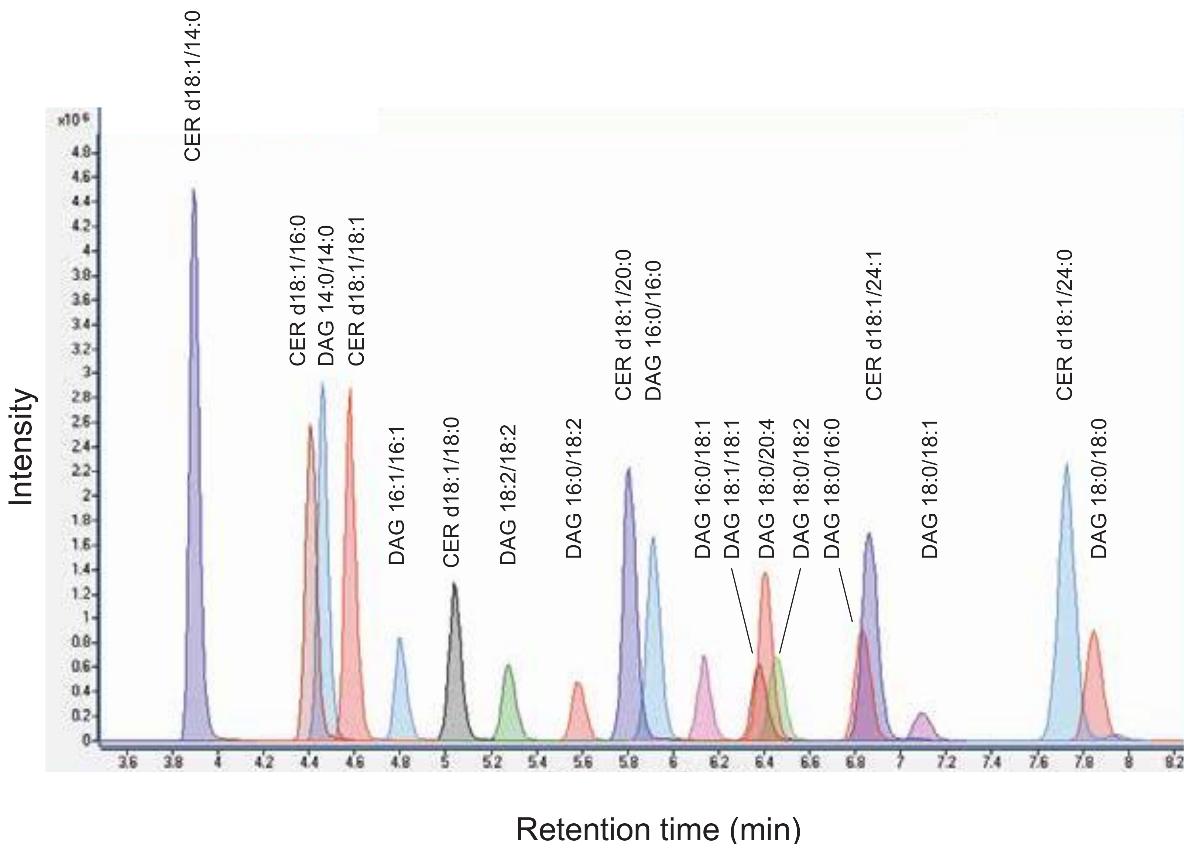
| Sample Type | Sample Collection | Lysis Method | Recommended Quantity | Additional Considerations |
|---|
| Tissues | Homogenization for cell release | Tissue-specific lysis protocols | 10-50 mg tissue | Tissue-specific considerations; avoid contamination during handling. |
| Cell Culture | Harvesting with trypsin or other methods | Specialized buffers for efficient lysis | Varies based on culture dish | Consistent culture conditions and avoiding contamination are crucial. |
| Plasma/Serum | Blood collection with anticoagulants | Protein precipitation or organic extraction | 100-500 μL | Immediate sample processing to prevent lipid alterations. |
| Cell Pellets | Centrifugation and washing | Specialized detergents for efficient lysis | 1-5 x 10^6 cells | Optimize washing steps to minimize cell residue. |
| Lipoproteins | Ultracentrifugation or density gradient | Organic extraction or specialized methods | 50-200 μL | Handle lipoproteins with care to prevent structural alterations. |
| Exosomes | Ultracentrifugation or precipitation | Specialized extraction methods | 100-500 μL | Maintain proper exosome isolation techniques for accurate analysis. |
| Breast Milk | Collection in clean containers | Lipid extraction from milk | 5-10 mL | Minimize contamination during collection and processing. |
| Skin Biopsy | Biopsy procedure | Tissue-specific lysis protocols | 5-10 mg tissue | Preserve sample integrity and prevent degradation. |
| Urine | Midstream collection in sterile containers | Organic extraction or precipitation | 5-10 mL | Process samples promptly to prevent lipid changes. |
| Feces | Fresh collection in airtight containers | Homogenization and organic extraction | 100-500 mg | Minimize oxygen exposure to maintain sample stability. |
| Plants | Harvest and flash freeze | Tissue-specific lysis protocols | 50-200 mg | Rapid freezing preserves lipid profiles in plant tissues. |
| Microorganisms | Culture or direct collection | Cell lysis and extraction methods | Varies based on biomass | Choose appropriate lysis methods for different microorganisms. |
| Food | Homogenization or extraction | Solvent-based extraction methods | Varies based on food type | Tailor extraction methods to the characteristics of the food sample. |