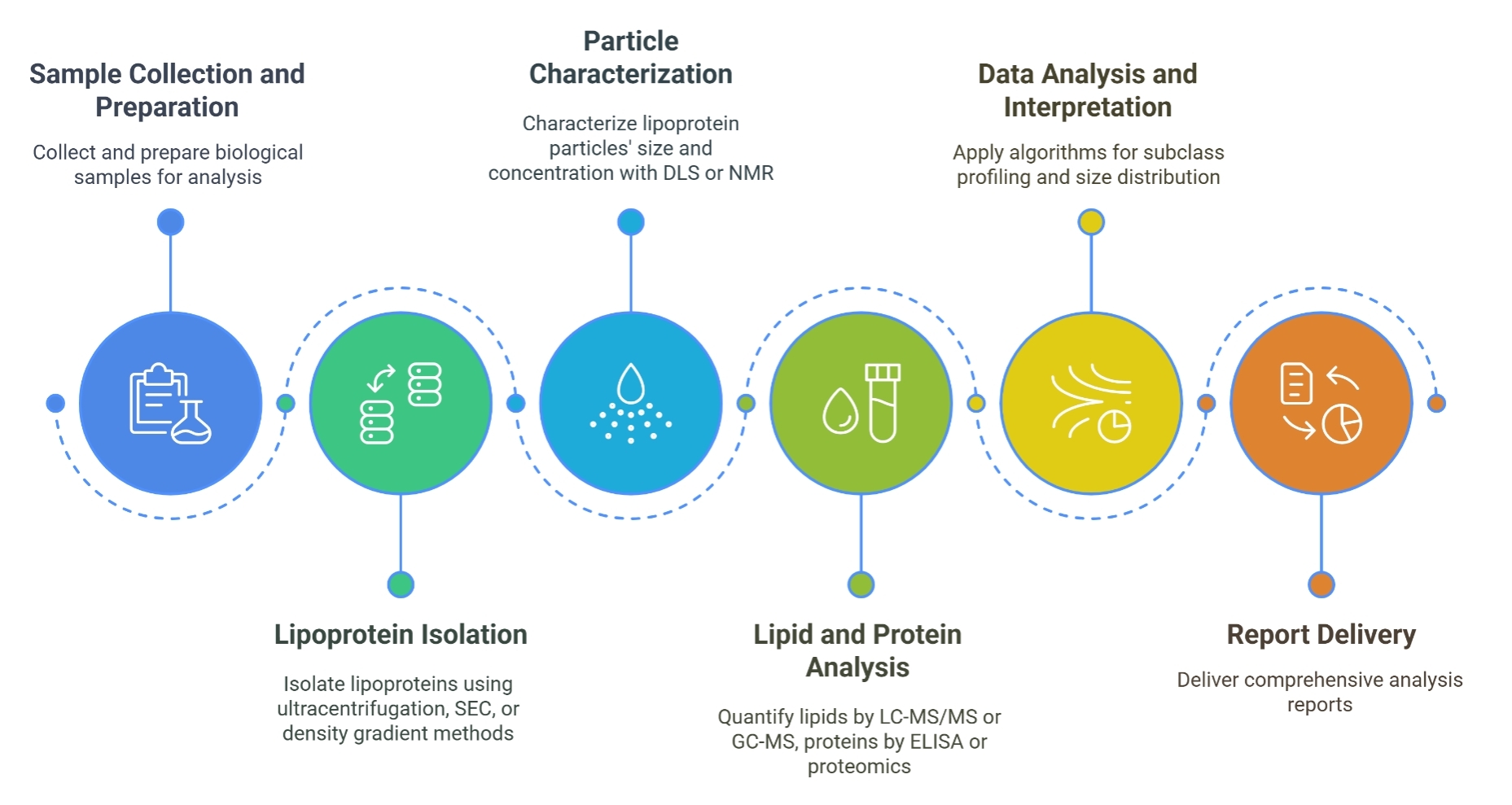
Lipoprotein Particle Profiling
Identification and quantification of different lipoprotein classes such as VLDL, LDL, HDL, and chylomicrons.
Our services have earned the trust of companies, schools, and organizations globally, and we remain dedicated to maintaining that trust.
Lipoproteins are particles composed of proteins and lipids. Their primary function is to carry hydrophobic lipids, such as cholesterol and triglycerides, throughout the bloodstream, making it possible for these substances to travel between organs and tissues that need them. Lipoproteins are classified into several types, each with specific roles in lipid metabolism and cardiovascular health. The major classes of lipoproteins include:
Analyzing lipoprotein levels and composition is vital for understanding lipid metabolism, cardiovascular disease risk, and metabolic disorders. Creative Proteomics specializes in providing comprehensive lipoprotein analysis that offers detailed insights into the lipid profile of biological samples.
Lipoprotein Particle Profiling
Identification and quantification of different lipoprotein classes such as VLDL, LDL, HDL, and chylomicrons.
Apolipoprotein Quantification
Measurement of specific apolipoproteins such as apoA-I, apoB, and apoE.
Lipidomic Profiling
Detailed analysis of the lipid composition within lipoprotein particles, including phospholipids, cholesterol, triglycerides, and fatty acids.
Lipoprotein Size and Density Analysis
Determination of the size distribution and density of lipoproteins using advanced techniques such as ultracentrifugation and gradient gel electrophoresis.
Quantification of Lipoprotein Subfractions
Measurement of lipoprotein subfractions, which may provide more precise information on lipid distribution and the associated cardiovascular risk.
| Lipoprotein Type | Size Range (nm) | Major Lipid Components | Key Proteins (Apolipoproteins) |
|---|---|---|---|
| Chylomicrons | 75–1,200 | Triglycerides, Cholesterol, Phospholipids | ApoB-48, ApoA-I, ApoC-II, ApoE |
| Very Low-Density Lipoprotein (VLDL) | 30–80 | Triglycerides, Cholesterol, Phospholipids | ApoB-100, ApoC-I, ApoC-II, ApoE |
| Intermediate-Density Lipoprotein (IDL) | 25–35 | Cholesterol, Triglycerides, Phospholipids | ApoB-100, ApoC-I, ApoE |
| Low-Density Lipoprotein (LDL) | 18–25 | Cholesterol, Phospholipids | ApoB-100 |
| High-Density Lipoprotein (HDL) | 5–15 | Cholesterol, Phospholipids | ApoA-I, ApoA-II, ApoC-I, ApoE |
| Lipoprotein(a) [Lp(a)] | 20–30 | Cholesterol, Phospholipids | ApoB-100, Apo(a) |
| Small Dense LDL | 18–22 | Cholesterol, Phospholipids | ApoB-100 |
| Large LDL | 22–25 | Cholesterol, Phospholipids | ApoB-100 |
| VLDL Remnants | 30–60 | Cholesterol, Triglycerides | ApoB-100, ApoC-I, ApoE |
| HDL2 | 8–12 | Cholesterol, Phospholipids | ApoA-I, ApoA-II |
| HDL3 | 5–8 | Cholesterol, Phospholipids | ApoA-I, ApoC-I |
| Triglyceride-Rich Lipoproteins | 20–80 | Triglycerides, Cholesterol, Phospholipids | ApoB-48, ApoB-100, ApoC-I, ApoC-II |
1. Ultracentrifugation and Gradient Density Separation
2. Size Exclusion Chromatography (SEC)
3. Mass Spectrometry (MS)
4. Nuclear Magnetic Resonance (NMR)
5. Dynamic Light Scattering (DLS)
Results Provided:
Format:
This report focuses on the analysis of apolipoproteins, key structural and functional components of lipoproteins.
Key Features:
This report details the lipidomic characteristics of lipoprotein particles, supporting comprehensive lipid metabolism assessments.
Key Features:
This report provides statistically driven insights to identify significant differences in lipoprotein profiles across experimental conditions.
Key Features:
Explore our Lipidomics Solutions brochure to learn more about our comprehensive lipidomics analysis platform.

Biomarker Identification in Basic Research
Identify and validate lipoprotein-related biomarkers for studying lipid metabolism regulation and its role in various biological processes.
Metabolic Research
Investigate lipid transport and metabolism pathways under various physiological and experimental conditions.

Toxicology Studies
Monitor lipoprotein changes as biomarkers of lipid metabolism disruption in response to chemical exposure or drug treatments.

Animal Model Research
Analyze lipoprotein profiles in preclinical animal models to understand lipid-related phenotypes and mechanisms.
Pharmaceutical Research and Development
Assess the effects of novel compounds on lipoprotein metabolism to support drug discovery and safety evaluation.

Nutritional and Dietary Studies
Evaluate the impact of specific diets, supplements, or nutritional interventions on lipid metabolism and lipoprotein composition.
| Sample Type | Minimum Volume | Storage Condition | Notes |
|---|---|---|---|
| Human Serum/Plasma | 45 µL | -80°C | Avoid hemolysis; use anticoagulants like EDTA. |
| Animal Serum/Plasma | 35 µL | -80°C | Suitable for mouse, rat, and other small animals. |
| Cerebrospinal Fluid (CSF) | 100 µL | -80°C | Ensure samples are clear without blood contamination. |
| Cell Culture Media | 500 µL | -80°C | Serum-free media preferred for accurate analysis. |
| Tissue Homogenates | 50 mg | -80°C | Flash freeze in liquid nitrogen; provide buffer details. |
| Lipoprotein Fractions | 100 µL | -80°C | Isolated via ultracentrifugation or SEC. |
| Other Biological Fluids | 200 µL | -80°C | Contact us for specific guidance. |
How do you ensure the accuracy of lipoprotein subclass profiling, especially for small particles like HDL3 or dense LDL?
We employ a combination of ultracentrifugation and size-exclusion chromatography (SEC) to achieve high-resolution separation of lipoprotein subclasses. For particles smaller than 10 nm (e.g., HDL3), we use dynamic light scattering (DLS) and nuclear magnetic resonance (NMR) to validate size distribution. Additionally, our mass spectrometry platform (Q Exactive™ HF) quantifies specific apolipoproteins and lipid components with a sensitivity of <0.1 ng/mL, ensuring precise subclass characterization even in low-abundance samples.
Can your service analyze oxidized or glycated lipoproteins, which are critical in studying atherosclerosis?
Yes. We offer targeted lipidomic workflows to detect oxidized phospholipids (e.g., oxLDL) and glycated apolipoproteins (e.g., glycated apoB) using high-resolution tandem mass spectrometry (HRMS/MS). For structural analysis, NMR spectroscopy identifies oxidation-induced conformational changes in lipoprotein particles. Customized protocols are available for studying post-translational modifications relevant to cardiovascular research.
What is the turnaround time for lipoprotein analysis, and can it be expedited for large-scale studies?
Standard turnaround time is 7–10 business days for routine profiling. For high-throughput projects (>500 samples), we implement automated batch processing with a guaranteed completion within 14 days. Expedited services (3–5 days) are available for urgent requests, leveraging our parallelized SEC-MS and robotic sample handling systems .
How do you handle species-specific differences in lipoprotein composition (e.g., mouse vs. human samples)?
Our platform includes species-specific reference databases for apolipoproteins and lipid profiles. For example:
What are the limitations of using serum vs. plasma for lipoprotein analysis?
While both serum and plasma are suitable, plasma (EDTA-treated) is preferred for:
How do you address matrix effects in complex samples like cerebrospinal fluid (CSF) or tissue homogenates?
For low-abundance samples (e.g., CSF), we use immunoaffinity depletion to remove high-abundance proteins (e.g., albumin) and enrich lipoproteins. Tissue homogenates undergo density gradient optimization to isolate intact particles. All methods are validated with spike-recovery experiments (85–115% recovery rate) .
Can your platform detect lipoprotein(a) [Lp(a)] and differentiate it from LDL?
Yes. Lp(a) is distinguished from LDL using agarose gel electrophoresis and apo(a)-specific antibodies in ELISA. Quantification via LC-MS/MS targets the unique kringle-IV domain of apo(a), avoiding cross-reactivity with apoB-100 .
How are samples protected from degradation?
All workflows include protease/phosphatase inhibitors and strict cold-chain logistics.
Do you provide raw data and bioinformatics support?
Raw files (mzML, .d) and statistical analysis (PCA, OPLS-DA) are included.
What is the detection limit for oxidized LDL?
<0.5 nM using our LC-MS/MS platform.

References
Services:
Resource:
Platform:
Online Inquiry
CONTACT US