Mycolic Acids Analysis Service
Creative Proteomics has developed a reliable and reproducible method using highly sensitive LC-MS/MS method for the rapid identification and quantification of Mycolic acids (MAs) in different sample types, which can satisfy the needs of academic and industrial study in your lab.
Overview
Mycolic acids (MAs) are major and specific lipid components of the mycobacterial cell envelope and are essential. They are found either unbound, extractable with organic solvents such as esters of trehalose or glycerol, or esterifying the terminal pentaarabinofuranosyl units of arabinogalactan (AG), the polysaccharide that, together with peptidoglycan, forms the insoluble cell wall skeleton. Both forms presumably play a crucial role in the remarkable architecture and impermeability of the cell envelope, participating in the two leaflets of the mycobacterial outer membrane, also called the mycomembrane.
Because the inhibition of MA synthesis is one of the main effects of the frontline and most efficient antitubercular drug INH, much attention has been paid to deciphering the chemistry and biosynthesis of MAs. The metabolic pathway of MAs represents a valuable source for recruiting latent targets for the development of novel anti-mycobacterial drugs in the alarming context of the emergence of extremely drug-resistant (XDR), multidrug-resistant (MDR), and totally drug-resistant (TDR) tuberculosis (TB). The last decade has seen impressive progress in the development of the biochemistry, genetics, regulation, and structures of MAs. Novel roles have also been attributed to MAs and their subfamilies in various phenomena, such as biofilm formation and foamy macrophage (FM) formation in TB granulomas.
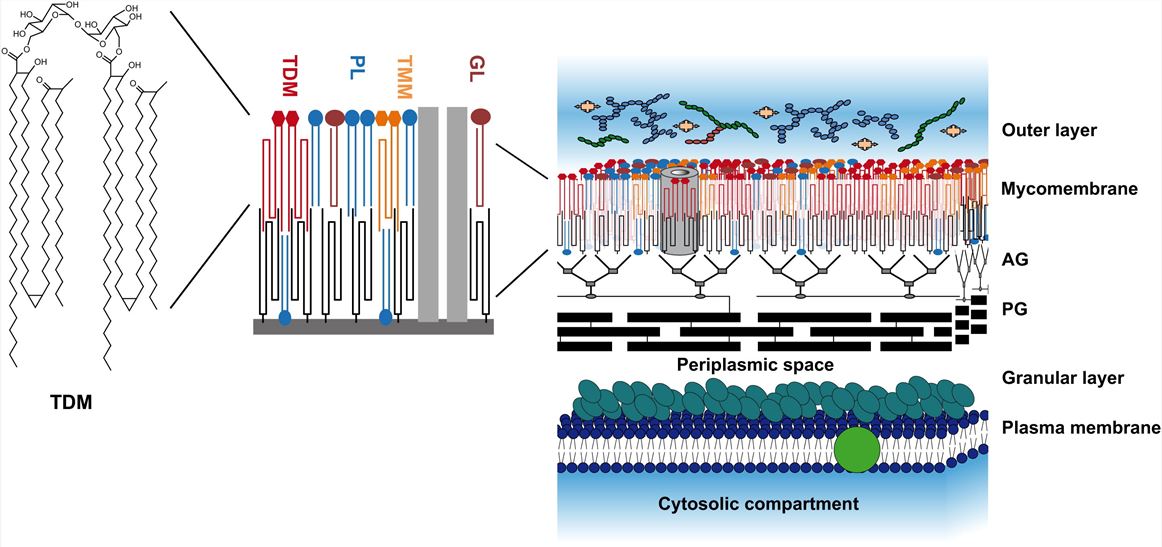 Fig1. Model of the Mycobacterial Cell Wall (Marrakchi, Hedia.; et al 2014)
Fig1. Model of the Mycobacterial Cell Wall (Marrakchi, Hedia.; et al 2014)
Currently, a reliable and reproducible method using highly sensitive LC-MS/MS platform for the identification and quantification of mycolic acids in different sample types has been established by the scientists at Creative Proteomics, which can satisfy the needs of academic and industrial study in your lab.
Why Choose Us?
- We use liquid chromatography hyphenated to triple quadrupole mass spectrometers for our MAs targeted lipidomics services, to provide high-specificity, high-selectivity, and high-reproducibility detection and quantitation of MAs.
- A detailed report will be provided at the end of the whole project, including the experiment procedure, MS/MS instrument parameter, raw data and data reports with figures, ready to be published.
- Our project is a very complex study that requires a high degree of sensitivity and accuracy. Creative Proteomics has molecular biologists, mass spectrometry specialists, bioinformatics experts, and lipidomics data scientists. We are well-prepared and positioned to foster collaboration in the fast-developing field of lipidomics.
Our Mycolic Acids Analysis Service
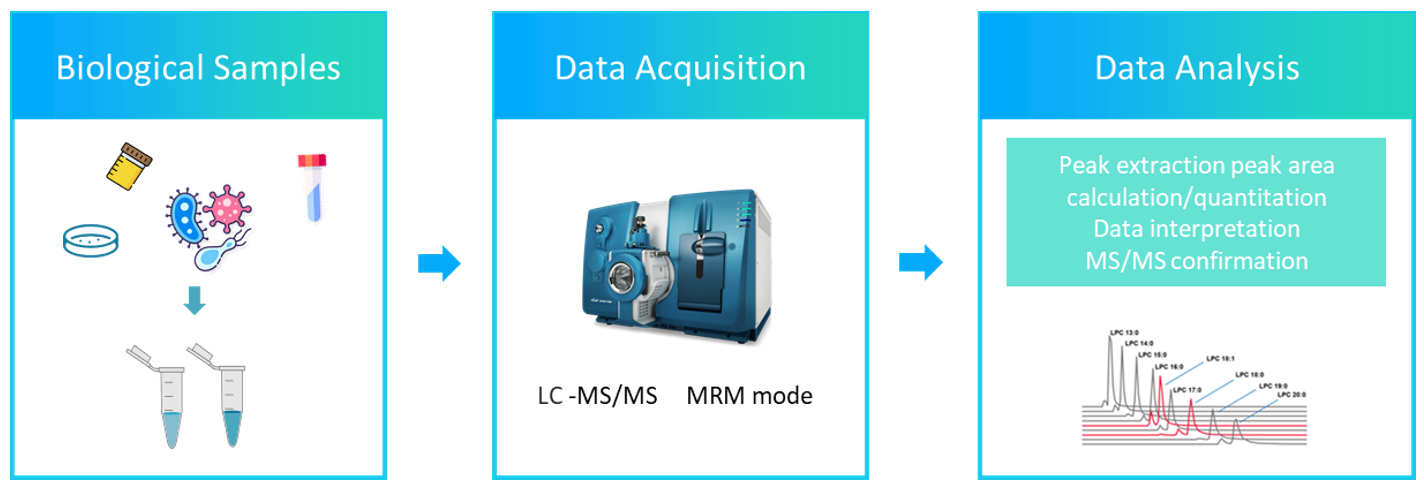 Fig 2. The workflow of mycolic acids analysis service
Fig 2. The workflow of mycolic acids analysis service
Structural details: species level
Mode: MRM
Precision: pmol & mol%
Delivery time: 2-4 weeks
Sample requirements
Blood/plasma: 100-200 ul/sample
Cells: 1x107/sample
Deliverables
- Experimental procedure
- Parameters of liquid chromatography and MS
- Purity analysis report
- MS raw data files and MS data quality checks
- Hexylceramide quantitative result data
- Custom analysis report
Based on a highly stable, reproducible and highly sensitive separation, characterization, identification and quantitative analysis system, combined with LC-MS/MS, Creative Proteomics provides reliable, fast and cost-effective mycolic acids analysis services. If you have any questions about our mycolic acids targeted lipidomics services, please contact us.
Reference:
- Marrakchi, Hedia.; et al. Mycolic acids: structures, biosynthesis, and beyond. Chemistry & biology. 2014, 21.1: 67-85.
* Our services can only be used for research purposes and Not for clinical use.
Services:
Resource:
Platform:


 Fig1. Model of the Mycobacterial Cell Wall (Marrakchi, Hedia.; et al 2014)
Fig1. Model of the Mycobacterial Cell Wall (Marrakchi, Hedia.; et al 2014) Fig 2. The workflow of mycolic acids analysis service
Fig 2. The workflow of mycolic acids analysis service