Our services have earned the trust of companies, schools, and organizations globally, and we remain dedicated to maintaining that trust.
What are Untargeted Lipidomics?
Untargeted Lipidomics is a high-throughput analytical approach used to study the lipid composition of biological samples without prior knowledge of the specific lipid species being analyzed. This untargeted method enables researchers to comprehensively profile all lipids present in a sample, identifying known and unknown lipid species alike.
In untargeted lipidomics, the goal is not only to detect and quantify lipids but also to capture the dynamic lipidome, revealing both the global lipid profile and specific lipidomic changes over time or in response to external stimuli. This approach is particularly valuable for discovering new lipid biomarkers and exploring novel therapeutic targets.
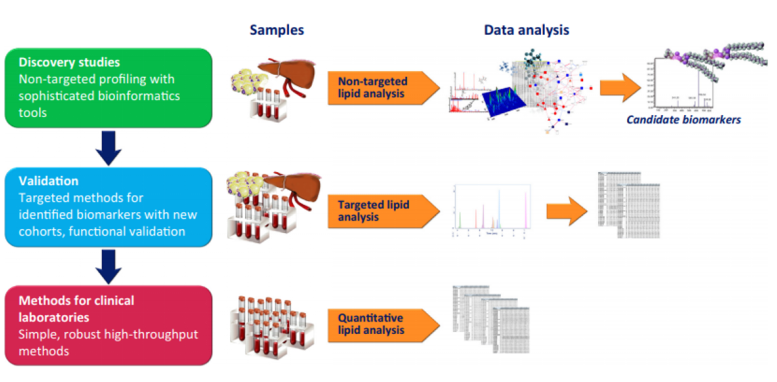
Lipidomics Profiling Analysis
At Creative Proteomics, our lipidomics profiling service uses advanced technologies and a refined workflow to ensure reliable and reproducible results. Our analytical equipment, including the Q Exactive HF and Q Exactive HF-X mass spectrometers, along with advanced data analysis software such as LipidSearch™, provide unmatched sensitivity, resolution, and precision for lipidomic studies.
Comprehensive Lipid Profiling
We offer a broad-spectrum analysis of lipids, covering a wide range of lipid classes, including but not limited to:
- Fatty Acyls (FA): Fatty acids, eicosanoids, etc.
- Glycerolipids (GL): Triglycerides, diglycerides, monoacylglycerols, etc.
- Glycerophospholipids (GP): Phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), etc.
- Sphingolipids (SL): Ceramides, sphingomyelin, glycosphingolipids, etc.
- Sterols (ST): Cholesterol, cholesterol esters, etc.
- Glycolipids (GL): Galactolipids (MGDG, DGDG), sulfolipids (SQDG), etc.
- Prenol Lipids (PL): Coenzyme Q, tocopherols, Vitamin K, etc.
Featured Untargeted Lipidomics Analysis
How Creative Proteomics Provides Lipidomics Profiling Analysis
What Platforms are Used for Our Untargeted Lipidomics Analysis?
Our lipidomics profiling service employs the latest mass spectrometry and chromatography technologies for precise and comprehensive lipid analysis.
Q Exactive HF/X Mass Spectrometers
The Q Exactive HF/X provides 10 ppm accuracy and Ultra High-Resolution (UHR) capabilities, ensuring highly sensitive and detailed lipid profiling. These instruments support both positive and negative ion mode for broad lipid coverage.
ACQUITY UPLC
We use ACQUITY UPLC CSH C18 columns for optimal separation of lipid species, allowing for accurate identification and quantification, even for complex lipid mixtures.
LipidSearch™ Software
Integrated with a lipid database of over 1.7 million lipid ions, LipidSearch™ software enables:
- Accurate lipid identification by matching detected peaks to database entries.
- Quantification of lipid levels across samples.
- Structural characterization of lipids, including fatty acid chain length and unsaturation.
ACQUITY UPLC (Figure from Waters)
Thermo Fisher Q Exactive (Figure from Thermo Fisher)
Comparing Shotgun and LC-MS for Untargeted Lipidomics Analysis
| Feature | Shotgun Untargeted Lipidomics | LC-MS Untargeted Lipidomics |
|---|
| Method | Direct mass spectrometry analysis without chromatography. | Lipid species are separated by liquid chromatography before MS analysis. |
| Analysis Speed | Faster due to no chromatography step. | Slower because of the chromatography separation. |
| Lipid Coverage | Broad, but may have lower resolution for complex lipids. | More detailed and higher resolution for complex lipids. |
| Sample Requirements | Requires less sample preparation, faster processing. | Requires more sample prep and longer analysis time. |
| Sensitivity | Moderate sensitivity; can struggle with complex samples. | Higher sensitivity due to chromatographic separation, ideal for low-abundance species. |
| Quantification Accuracy | Lower accuracy in quantification for complex mixtures. | Higher accuracy in quantification, especially for co-eluting species. |
| Ion Interference | Higher potential for ion suppression and interference. | Lower ion interference due to better separation. |
| Resolution | Lower resolution for complex samples (ion overlap possible). | Higher resolution due to chromatographic separation. |
| Throughput | Higher throughput due to simplified workflow. | Lower throughput but better suited for detailed studies. |
| Ideal For | Large-scale, high-throughput lipid screening with minimal sample. | In-depth lipid profiling and complex mixture analysis where high resolution and accuracy are needed. |
Why Choose Our Untargeted Lipidomics Services?
- Broad Lipid Coverage: We identify and quantify 100+ lipid classes and over 4,200 individual lipids, from storage lipids to membrane and signaling lipids, ensuring comprehensive lipid profiling.
- Minimal Sample Requirements: Our analysis needs only 1 µL of blood plasma to deliver accurate lipid data, allowing work with small or limited samples.
- High Sensitivity and Resolution: With Q Exactive HF/X mass spectrometers, we achieve <1 ppm mass accuracy and sub-femtomole sensitivity for detecting low-abundance lipids.
- Fast Turnaround: Our high-throughput workflow processes thousands of samples weekly, delivering results in as little as 2 weeks, accelerating your research.
- Versatility Across Sample Types: We analyze a wide range of biological samples, including tissues, fluids, cells, bacteria, and more, supporting diverse research needs.
- Tailored Project Design: We offer customized recommendations for lipidomics platforms, sample prep, and lipid extraction to suit your specific study.
- High-Throughput Capabilities: We process hundreds to thousands of samples per week, ideal for large-scale and high-throughput research.
- Discovery of Novel Biomarkers: Untargeted lipidomics uncovers novel lipids, with up to 30% of detected species being new, enabling fresh insights into disease mechanisms.
- Lipid Metabolism Insights: We provide a global view of lipid metabolism, highlighting key alterations in >30 metabolic pathways, crucial for understanding diseases.
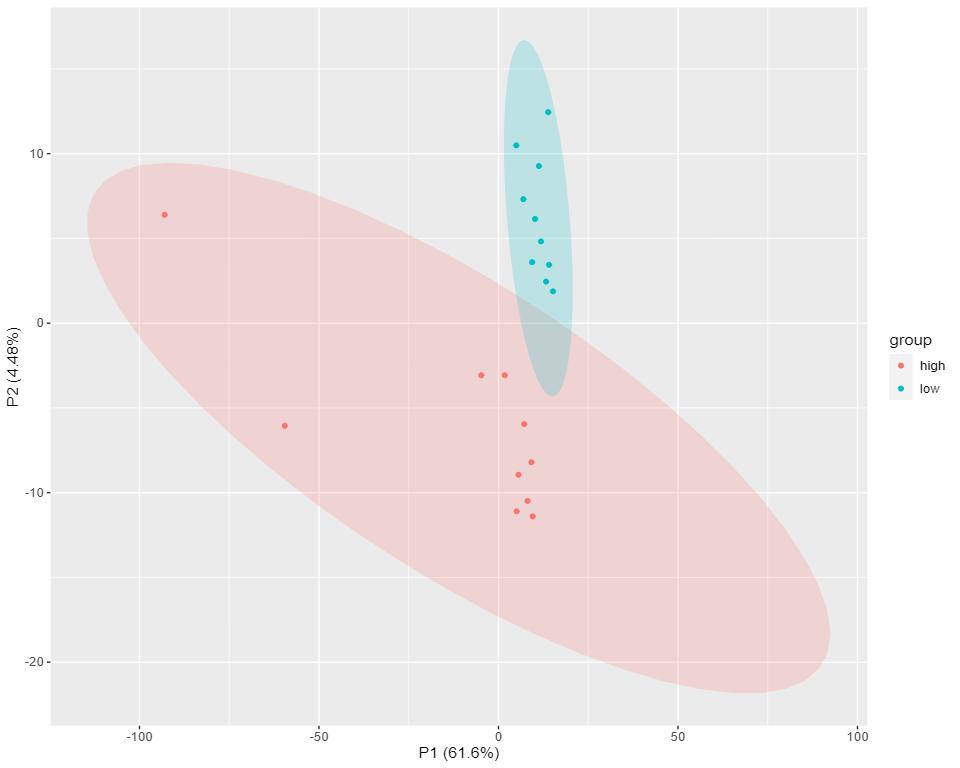
- Robust Data Analysis: Advanced tools like PCA and PLS-DA ensure statistically significant results (p-value < 0.05), identifying differential lipids and biomarkers.
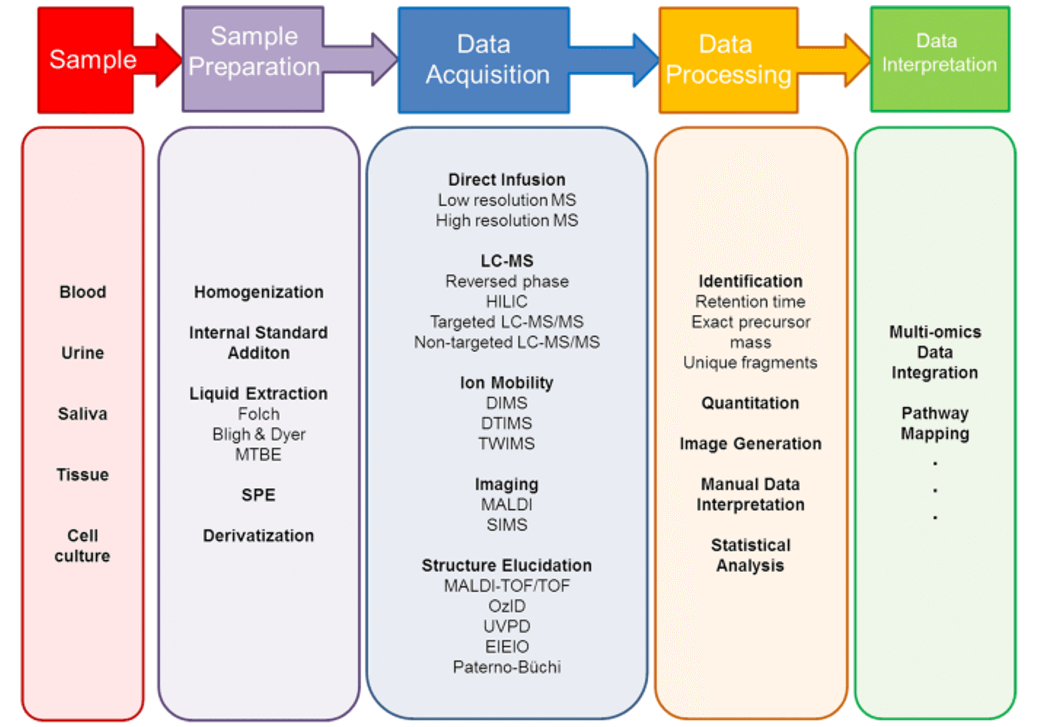
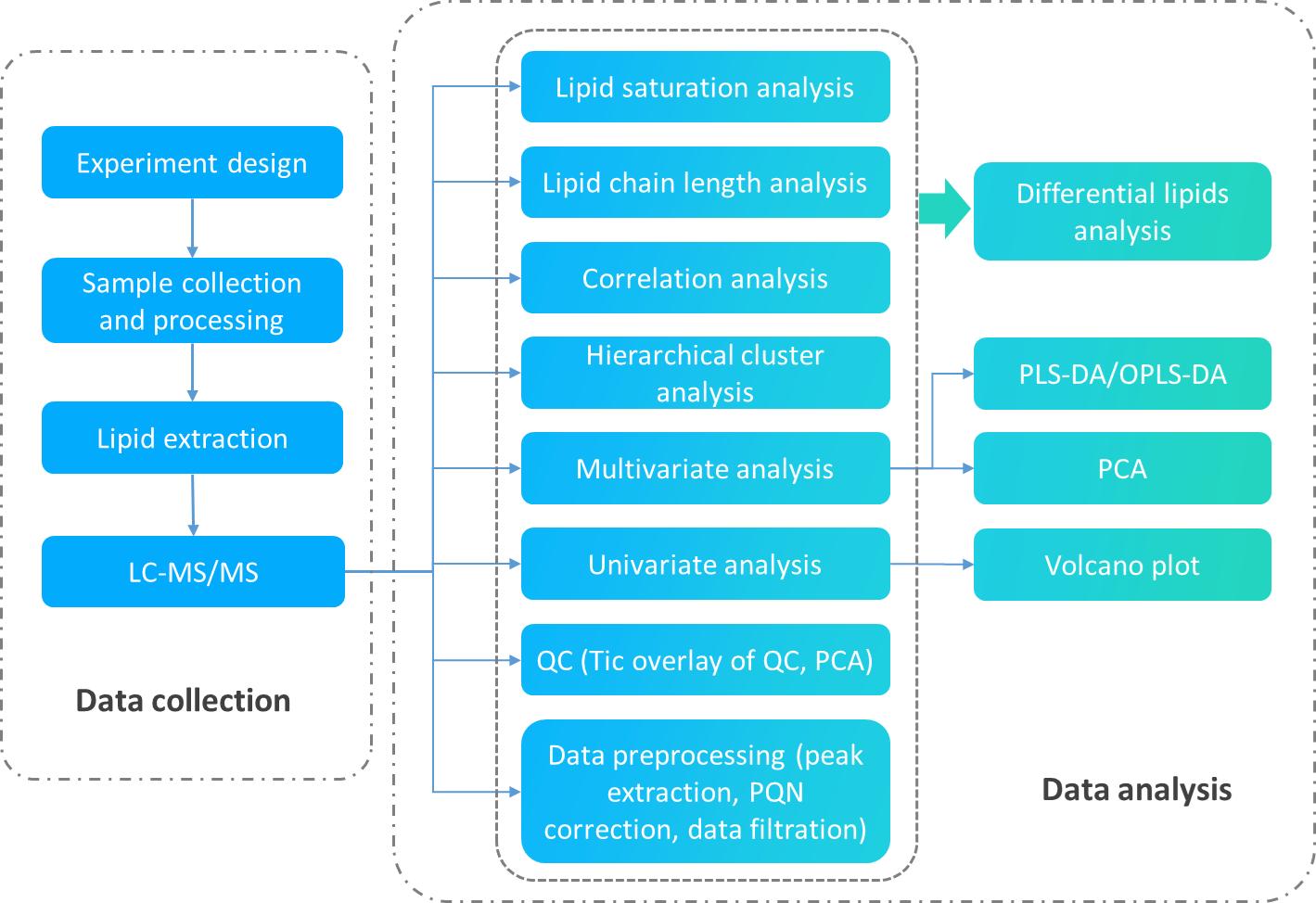
Untargeted Lipidomics Analysis Service: Results and Data Analysis
Lipid Species Identification: Accurate identification of lipid species based on mass spectral data and comparison to extensive lipid databases.
Quantification of Lipids: Quantification of lipid species across multiple samples, enabling comparison of lipid levels in different conditions (e.g., disease vs. healthy).
Lipid Class Profiling: Detailed profiling of lipid classes such as phospholipids, triglycerides, sphingolipids, fatty acids, and others.
Lipid Distribution Across Samples: Analysis of lipid distribution within biological samples (e.g., tissues, cells, plasma) to uncover patterns of lipid metabolism.
Metabolite Pathway Analysis: Mapping of lipid species to known metabolic pathways, providing insight into lipid metabolism and signaling processes.
Differential Lipid Expression: Identification of differentially expressed lipids across experimental conditions (e.g., control vs. treatment, disease vs. normal).
Structural Characterization: Structural characterization of lipids, including head group type, fatty acid chain length, and unsaturation levels.
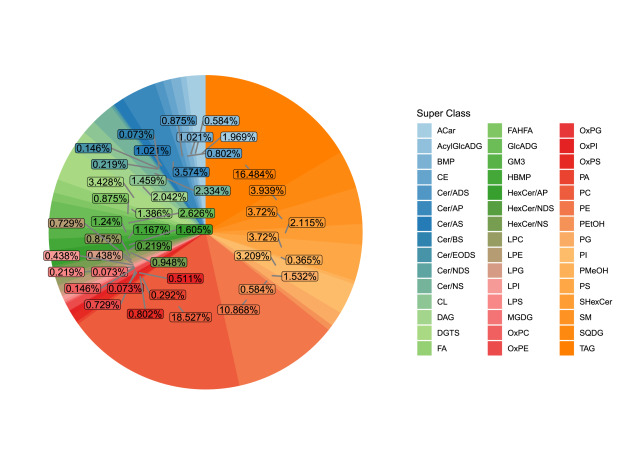
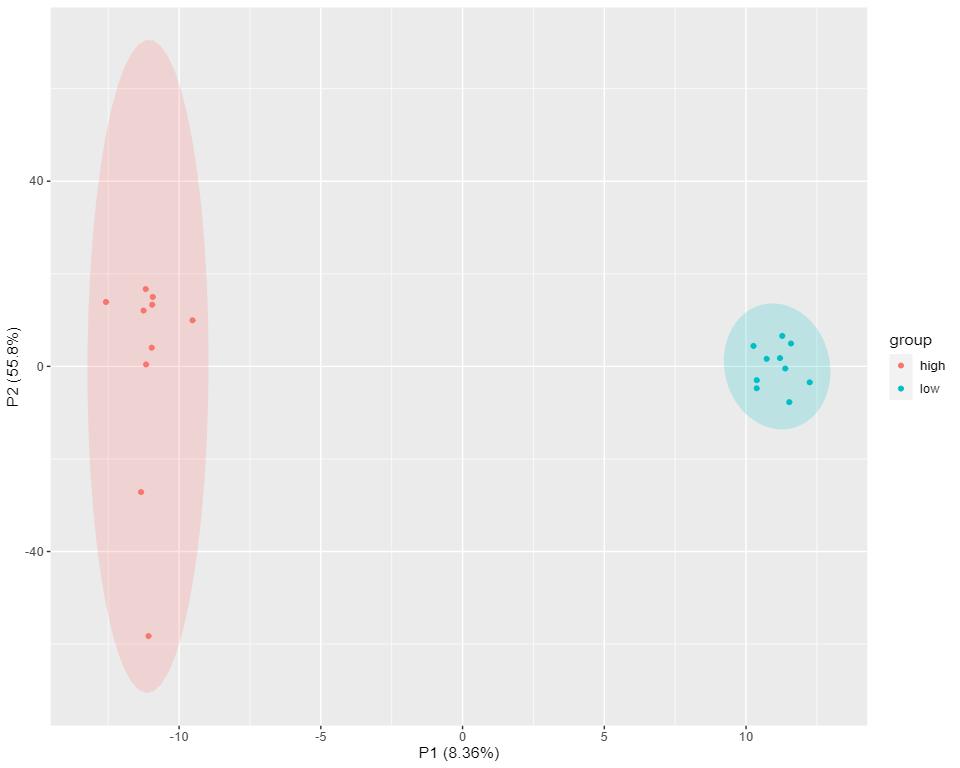
Pie chart of distribution of detected complex lipid subclasses.
Figure from Yang Biochimie 2023.
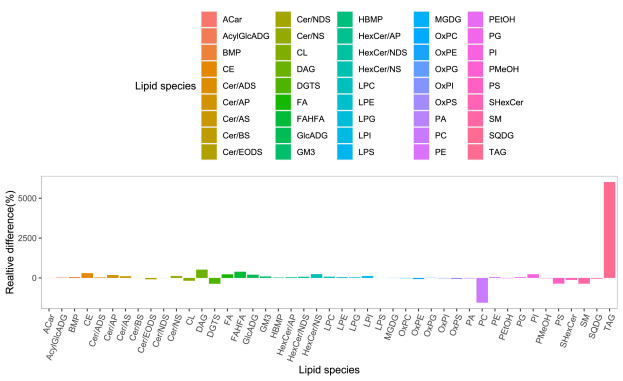
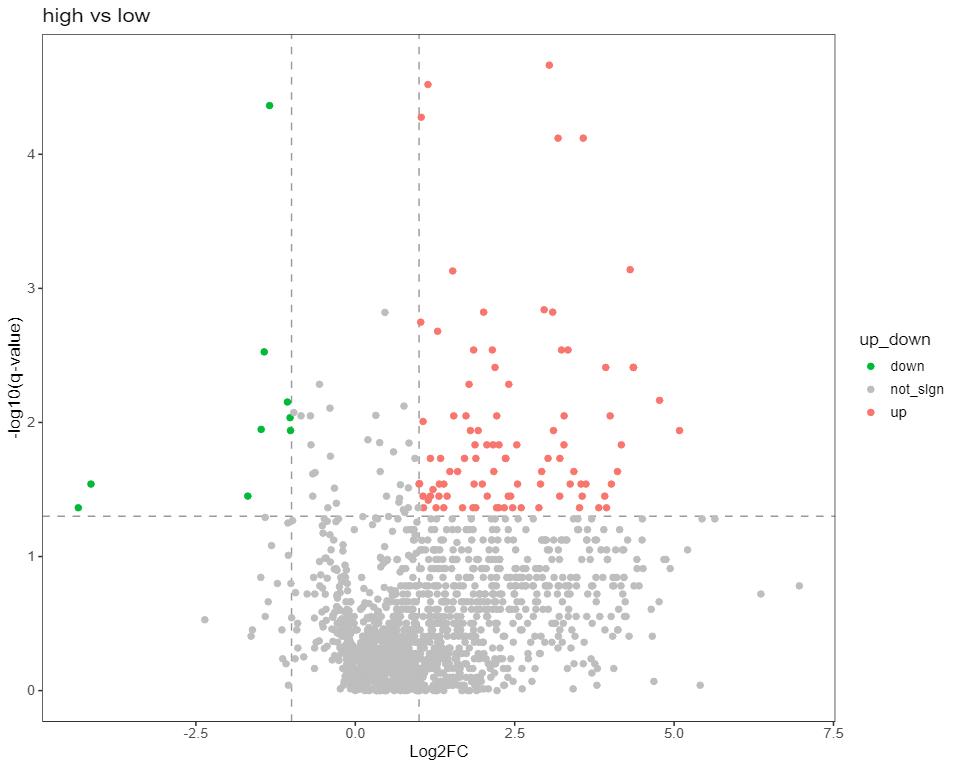
The bar plot shows the relative difference in the detected significantly different lipids
Figure from Yang Biochimie 2023.
Explore our Lipidomics Solutions brochure to learn more about our comprehensive lipidomics analysis platform.
Download Brochure
What Our Untargeted Lipidomics Analysis Used For
In the pharmaceutical and biotechnology fields, lipidomics is used to identify lipid-based biomarkers, study the effects of drugs, and understand the underlying mechanisms of diseases. By analyzing how lipids change in response to treatments, researchers can improve drug development and discover new therapeutic targets.
Lipidomics is vital in food science, helping to analyze the lipid content of different foods and their effects on health. It provides valuable insights into how dietary fats impact metabolic processes, inflammation, and the risk of chronic diseases like obesity and heart disease.
Lipidomics is increasingly used in disease research to explore how lipids contribute to conditions like cancer, cardiovascular diseases, neurodegeneration, and metabolic disorders. By examining lipid profiles in blood, tissues, and cells, researchers can identify new biomarkers and potential treatments.
In environmental toxicology, lipidomics helps assess how environmental pollutants affect lipid metabolism in animals, plants, and humans. It provides insights into how exposure to chemicals like pesticides or industrial toxins can alter lipid profiles, helping to identify potential health risks and environmental hazards.
Lipidomics can be applied to agriculture to study how plants and animals use lipids for growth, energy storage, and stress response. It helps improve crop quality, enhance resistance to diseases, and optimize animal health, supporting better agricultural practices and food production.
In microbiology, lipidomics is used to analyze the lipid profiles of bacteria, fungi, and other microorganisms. This research helps understand how microorganisms adapt to different environments, resist stress, or cause infections, and can lead to new ways of combating antibiotic resistance or improving industrial fermentation processes.
Sample Requirements for Untargeted Lipidomics Analysis Solutions
| Sample Type | Recommended Sample Amount | Minimum Sample Amount | Notes |
|---|
| Serum / Plasma | ≥ 300 µL | ≥ 100 µL | Fresh or frozen samples, stored at -80°C until analysis. |
| Urine | ≥ 300 µL | ≥ 100 µL | Fresh samples preferred; use sterile containers. |
| Saliva / Seminal Plasma / Milk | ≥ 300 µL | ≥ 100 µL | Store at -80°C for long-term storage. |
| Animal Tissue (e.g., liver, brain) | ≥ 200 mg | ≥ 25 mg | Homogenized and stored at -80°C. |
| Plant Tissue (e.g., roots, leaves) | ≥ 200 mg | ≥ 25 mg | Samples should be quickly frozen to avoid lipid degradation. |
| Feces / Intestinal Contents | ≥ 200 mg | ≥ 25 mg | Use sterile tubes for collection, store at -80°C. |
| Suspension Cells (e.g., cultured cells) | ≥ 10⁷ cells | ≥ 5 × 10⁶ cells | Cells should be harvested at peak growth phase. |
| Adherent Cells (e.g., tissue culture) | ≥ 10⁷ cells | ≥ 5 × 10⁶ cells | Cells should be washed to remove any culture medium. |
| Microorganisms | ≥ 200 mg | ≥ 25 mg | For bacteria or fungi, collect in sterile conditions. |
| Culture Medium / Fermentation Medium | ≥ 1 mL | ≥ 100 µL | Collect media free of cells or contaminants. |
| Cerebrospinal Fluid (CSF) | ≥ 300 µL | ≥ 100 µL | Store at -80°C; handle under sterile conditions. |
| Amniotic Fluid / Hemolymph | ≥ 300 µL | ≥ 100 µL | Ensure sample is collected under sterile conditions. |
FAQs for Untargeted Lipidomics Analysis Service
Can I use formalin-fixed samples for untargeted lipidomics analysis?
It's generally not recommended to use formalin-fixed samples for lipidomics, as formalin fixation can alter or cross-link lipid molecules, making them harder to extract and analyze. We recommend using fresh or snap-frozen samples to ensure accurate lipid profiling.
What is the difference between untargeted lipidomics and targeted lipidomics?
In untargeted lipidomics, the goal is to identify and quantify as many lipid species as possible, without predefined biases. This allows for the discovery of novel lipid markers or metabolic pathways associated with diseases. In contrast, targeted lipidomics focuses on the quantification of specific lipid species or classes, typically chosen based on prior knowledge or research hypotheses. While untargeted lipidomics offers a broader overview, targeted lipidomics provides more accurate and reproducible quantification of specific lipids.
How many biological replicates do I need for an untargeted lipidomics study?
For robust data and meaningful conclusions, we recommend using at least 3-5 biological replicates per experimental condition. The number of replicates helps ensure that your results are statistically significant and reduces the risk of false positives or over-interpretation of data from outliers. However, more replicates may be necessary for complex experimental designs or when working with small sample sizes.
What chromatographic methods are most used in lipidomics?
Reversed-phase liquid chromatography (RP-LC) is commonly used for non-polar lipids, while normal-phase chromatography (NP-LC) works better for polar lipids like phospholipids.
What is the role of internal standards in lipidomics?
Internal standards, often stable isotope-labeled lipids, correct for sample variability and improve quantitative accuracy, even in untargeted analysis.
Reference
- Yang, Yu, et al. "LC-MS/MS based untargeted lipidomics uncovers lipid signatures of late-onset preeclampsia." Biochimie 208 (2023): 46-55.