What are Phospholipids?
Phospholipids are a class of lipids that are a crucial component of the cell membrane. Structurally, these molecules are characterized by a hydrophilic "head" that contains a phosphate group, and two hydrophobic "tails" composed of fatty acids. Due to this unique structure, phospholipids serve as the primary building block for cell membranes, where they provide critical functions, including compartmentalization, protein trafficking, and signal transduction.
Phospholipids Analysis Service Projects by Creative Proteomics
Creative Proteomics offers a wide range of specialized services for the analysis of phospholipids, tailored to meet the diverse needs of our clients. Our comprehensive portfolio includes:
- Phospholipid Profiling: Comprehensive profiling of phospholipid species in biological samples, providing insights into lipid composition and distribution.
- Phospholipid Quantification: Accurate quantification of phospholipids to determine their abundance and concentration levels in various biological matrices.
- Phospholipid Structural Elucidation: Detailed characterization of phospholipid molecular structures, including identification of fatty acid chains and headgroup compositions.
- Phospholipid Metabolite Analysis: Investigation of phospholipid metabolites and their role in metabolic pathways and disease mechanisms.
- Phospholipid Biomarker Discovery: Identification of phospholipid biomarkers associated with disease states or physiological conditions for diagnostic and therapeutic applications.
- Phospholipid Interaction Studies: Assessment of phospholipid interactions with proteins, small molecules, or other lipids to elucidate their functional roles in cellular processes.
- Phospholipid Stability Studies: Evaluation of phospholipid stability under different storage conditions or environmental factors to optimize sample handling protocols.
- Customized Phospholipid Analysis: Tailored services to meet specific research needs or project requirements, including method development, validation, and optimization.
Detected Phospholipid Species
Advanced Technologies for Phospholipids Analysis
Liquid Chromatography-Mass Spectrometry (LC-MS)
Liquid chromatography-mass spectrometry (LC-MS) serves as the cornerstone of our phospholipids analysis service. LC-MS offers exceptional sensitivity, selectivity, and resolution, enabling precise identification and quantification of phospholipid species in complex biological matrices. By coupling high-performance liquid chromatography (HPLC) with mass spectrometry, we achieve efficient separation of phospholipids based on their chemical properties and molecular weights. Electrospray ionization (ESI) facilitates the ionization of non-volatile phospholipids, allowing for direct analysis without the need for sample derivatization. Our LC-MS platform provides comprehensive coverage of phospholipid classes and molecular species, facilitating in-depth lipidomic profiling and structural elucidation.
Ultra-performance Liquid Chromatography (UPLC)
UPLC is an essential component of our phospholipids analysis platform, enabling the efficient separation of complex phospholipid mixtures with superior resolution compared to traditional liquid chromatography methods. By employing a combination of UPLC and reverse-phase liquid chromatography, we ensure precise separation and detection of a wide range of phospholipid species, including phosphatidylcholine, phosphatidylinositol, and phosphatidylserine. UPLC enhances our ability to characterize phospholipid profiles in biological samples with unparalleled accuracy and efficiency.
High-Resolution Mass Spectrometry (HRMS)
To provide detailed insights into the composition and structure of phospholipids, Creative Proteomics incorporates High-Resolution Mass Spectrometry into its phospholipids analysis service. HRMS offers a high level of accuracy, allowing for detailed identification and quantification of phospholipid species. It directly measures the mass-to-charge ratio of ionized phospholipids, providing accurate information about their molecular structure. This technology enhances our ability to elucidate the complex molecular makeup of phospholipids and their role in biological systems.
Gas Chromatography-Mass Spectrometry (GC-MS)
GC-MS complements our LC-MS capabilities for specific phospholipid analyses, particularly for the characterization of phospholipid fatty acid composition following derivatization. By converting phospholipid fatty acids into volatile derivatives, GC-MS enables the separation and quantification of individual fatty acid species. This technique is invaluable for studying lipid metabolism, membrane dynamics, and lipidomic alterations in disease states.
Multi-reaction Monitoring (MRM)
In conjunction with UPLC and HRMS, Creative Proteomics uses Multi-reaction Monitoring to improve the sensitivity and selectivity of phospholipids analysis. MRM enables the detection and quantification of low-abundance phospholipids, even in complex biological samples. It serves as a complementary tool to HRMS, providing validation of results and enhancing confidence in data accuracy.
Nuclear Magnetic Resonance (NMR) Spectroscopy
NMR spectroscopy offers unique insights into phospholipid structure, dynamics, and interactions. Our NMR platform enables non-destructive analysis of phospholipids in solution, providing information on molecular conformation, lipid-lipid interactions, and membrane organization. By probing phospholipid headgroup and acyl chain dynamics, NMR spectroscopy contributes to our understanding of membrane biophysics and lipid-mediated cellular processes.
Integration of Technologies
By integrating these advanced technologies, Creative Proteomics provides a comprehensive platform for phospholipids analysis service. This integrated approach enables the separation, detection, and quantification of a wide variety of phospholipid molecules in biological samples, offering detailed insights into their role in various physiological and pathological processes. Our advanced technology platform is dedicated to supporting research endeavors and facilitating breakthroughs in the biomedical field.
Sample Requirements for Phospholipids Analysis
| Sample Type | Sample Volume | Storage Conditions | Additional Notes |
|---|
| Biological Samples | Minimum: 50 µL | -80°C or below | Avoid repeated freeze-thaw cycles. |
| Tissue Homogenates | Minimum: 20 mg | -80°C or below | Homogenize in appropriate buffer or solvent. |
| Cell Culture Media | Minimum: 100 µL | -80°C or below | Centrifuge to remove debris before storage. |
| Plasma/Serum | Minimum: 100 µL | -80°C or below | Ensure samples are collected using appropriate anticoagulants. |
If you have any questions about our lipidomics services, please contact us.
Case Comprehensive Analysis of Phospholipids Using HPLC-MS/MS
Background
Phospholipids are crucial components of biological membranes, playing vital roles in various cellular processes. However, their structural diversity, including isobaric and isomeric species, presents challenges for accurate characterization. Traditional methods, such as MALDI-TOF analysis, may lead to misinterpretations due to the complexity of phospholipid mixtures. Advanced techniques like HPLC-MS/MS offer improved resolution and specificity, facilitating in-depth phospholipid analysis.
Samples
Bovine heart mitochondria (BHM) samples were utilized for phospholipid analysis. These samples are particularly relevant for studying mitochondrial membranes, where phospholipids play essential roles in membrane structure and function.
Technical Methods
Sample Preparation:
- Phospholipids were extracted from bovine heart mitochondria (BHMs), a source rich in phospholipids.
- Extraction methods likely involved organic solvents such as chloroform/methanol, known for their efficiency in extracting lipids from biological samples.
Instrumentation:
- High-performance liquid chromatography (HPLC) coupled with tandem mass spectrometry (MS/MS) was employed.
- The HPLC system likely utilized a normal-phase column, which separates compounds based on differences in polarity.
- Tandem mass spectrometry (MS/MS) involved the use of a mass spectrometer equipped with collision-induced dissociation (CID) capabilities.
Chromatographic Separation:
- The chromatographic separation was crucial for resolving different classes of phospholipids and individual species within each class.
- Normal-phase chromatography is preferred for separating phospholipids based on their polar headgroups and acyl chain composition.
Mass Spectrometry Analysis:
- Phospholipid ions were introduced into the mass spectrometer, likely via electrospray ionization (ESI) in negative ion mode, which is commonly used for analyzing phospholipids.
- Collision-induced dissociation (CID) was employed to fragment phospholipid ions in the gas phase.
- Fragment ions generated during CID were analyzed to deduce the structural characteristics of phospholipids, such as acyl chain composition and sn-positioning.
Data Analysis:
- Data analysis likely involved matching observed fragment ions with theoretical fragmentation patterns of phospholipids.
- The identification and structural characterization of phospholipids were based on the presence and relative intensities of characteristic fragment ions in the MS/MS spectra.
- The interpretation of MS/MS spectra required expertise in lipid chemistry and mass spectrometry.
Verification and Confirmation:
- The proposed structures of phospholipid species were likely verified through comparison with standard compounds and additional experiments, such as MS3 characterization of certain ions.
- The robustness and reliability of the analytical method were validated through reproducibility experiments and comparison with previous studies.
Quantification (Possibly):
- Quantitative analysis of phospholipid species may have been performed using internal standards or calibration curves, although specific details on quantification methods are not provided in the summary.
Results
Challenges in CL Characterization: CL, with its dimeric structure, presented significant challenges due to numerous isomeric species. The study identified CL species containing odd-numbered acyl chains, a novel finding in mammalian mitochondria.
Characterization of MLCL and DLCL: Despite their low abundance, MLCL and DLCL species were successfully characterized using MS/MS peaks corresponding to MAG-like and DAG-like ions, highlighting the efficacy of HPLC-MS/MS.
Complexities in Other Phospholipids: PE, PC, PG, PI, and PS exhibited complexities due to numerous isomeric and isobaric species. Correcting misassignments from previous analyses, the study identified certain species as ether-linked PE instead of PS.
Advantages of HPLC Separation: HPLC separation facilitated not only class separation but also the identification of minor species, enhancing the accuracy and comprehensiveness of phospholipid characterization.
Consistent Observations Across Phospholipid Classes: More saturated fatty acids were consistently found at the sn-1 position across phospholipid classes, supporting previous structural observations and highlighting the utility of MAG-like ions in determining acyl chain positions.
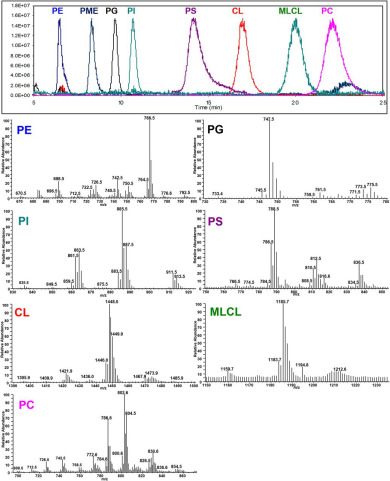 Normalized ion chromatograms of PE, PG, PI, PS, CL, MLCL, and PE from BHMs and PME, the internal standard (upper panel) and mass spectra of individual classes of phospholipids (lower panels).
Normalized ion chromatograms of PE, PG, PI, PS, CL, MLCL, and PE from BHMs and PME, the internal standard (upper panel) and mass spectra of individual classes of phospholipids (lower panels).
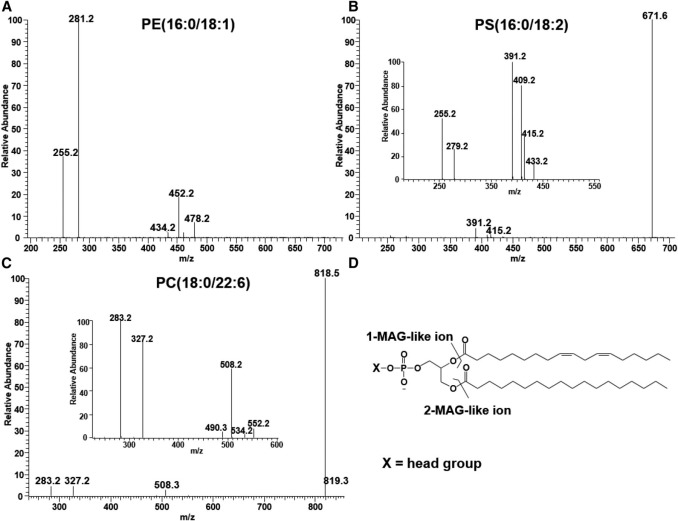 The MS/MS spectra of standard synthetic phospholipids PE(16:0/18:1), m/z 716.5 (A), PS(16:0/18:2), m/z 758.5 (B), and PC(18:0/22:6), m/z 878.5 (C), obtained in the negative ion mode, and fragmentation of phospholipids leading to1-MAG-like ions and 2-MAG-like ions (D).
The MS/MS spectra of standard synthetic phospholipids PE(16:0/18:1), m/z 716.5 (A), PS(16:0/18:2), m/z 758.5 (B), and PC(18:0/22:6), m/z 878.5 (C), obtained in the negative ion mode, and fragmentation of phospholipids leading to1-MAG-like ions and 2-MAG-like ions (D).
![The MS/MS spectrum of PG molecular species resulting after collisional activation of the [M-H]− ions at m/z 747.5 (A) and 733.5 (B).](upload/image/phospholipids-analysis-service-3.jpg) The MS/MS spectrum of PG molecular species resulting after collisional activation of the [M-H]− ions at m/z 747.5 (A) and 733.5 (B).
The MS/MS spectrum of PG molecular species resulting after collisional activation of the [M-H]− ions at m/z 747.5 (A) and 733.5 (B).
![The MS/ MS spectrum of PI molecular species resulting after collisional activation of the [M-H]− ions at m/z 885.5 (A) and 835.5 (B).](upload/image/phospholipids-analysis-service-4.jpg) The MS/ MS spectrum of PI molecular species resulting after collisional activation of the [M-H]− ions at m/z 885.5 (A) and 835.5 (B).
The MS/ MS spectrum of PI molecular species resulting after collisional activation of the [M-H]− ions at m/z 885.5 (A) and 835.5 (B).
![The MS/MS spectrum of PS molecular species resulting after collisional activation of the [M-H]− ions at m/z 794.5 (A) and 772.5 (B).](upload/image/phospholipids-analysis-service-5.jpg) The MS/MS spectrum of PS molecular species resulting after collisional activation of the [M-H]− ions at m/z 794.5 (A) and 772.5 (B).
The MS/MS spectrum of PS molecular species resulting after collisional activation of the [M-H]− ions at m/z 794.5 (A) and 772.5 (B).
![The MS/ MS spectrum of PC molecular species resulting after collisional activation of the [M+HCOO]− ions at m/z 802.5 (A) and 786.5 (B)](upload/image/phospholipids-analysis-service-6.jpg) The MS/ MS spectrum of PC molecular species resulting after collisional activation of the [M+HCOO]− ions at m/z 802.5 (A) and 786.5 (B)
The MS/ MS spectrum of PC molecular species resulting after collisional activation of the [M+HCOO]− ions at m/z 802.5 (A) and 786.5 (B)
Reference
- Kim, Junhwan, and Charles L. Hoppel. "Identification of unusual phospholipids from bovine heart mitochondria by HPLC-MS/MS." Journal of lipid research 61.12 (2020): 1707-1719.


 Normalized ion chromatograms of PE, PG, PI, PS, CL, MLCL, and PE from BHMs and PME, the internal standard (upper panel) and mass spectra of individual classes of phospholipids (lower panels).
Normalized ion chromatograms of PE, PG, PI, PS, CL, MLCL, and PE from BHMs and PME, the internal standard (upper panel) and mass spectra of individual classes of phospholipids (lower panels). The MS/MS spectra of standard synthetic phospholipids PE(16:0/18:1), m/z 716.5 (A), PS(16:0/18:2), m/z 758.5 (B), and PC(18:0/22:6), m/z 878.5 (C), obtained in the negative ion mode, and fragmentation of phospholipids leading to1-MAG-like ions and 2-MAG-like ions (D).
The MS/MS spectra of standard synthetic phospholipids PE(16:0/18:1), m/z 716.5 (A), PS(16:0/18:2), m/z 758.5 (B), and PC(18:0/22:6), m/z 878.5 (C), obtained in the negative ion mode, and fragmentation of phospholipids leading to1-MAG-like ions and 2-MAG-like ions (D).![The MS/MS spectrum of PG molecular species resulting after collisional activation of the [M-H]− ions at m/z 747.5 (A) and 733.5 (B).](upload/image/phospholipids-analysis-service-3.jpg) The MS/MS spectrum of PG molecular species resulting after collisional activation of the [M-H]− ions at m/z 747.5 (A) and 733.5 (B).
The MS/MS spectrum of PG molecular species resulting after collisional activation of the [M-H]− ions at m/z 747.5 (A) and 733.5 (B).![The MS/ MS spectrum of PI molecular species resulting after collisional activation of the [M-H]− ions at m/z 885.5 (A) and 835.5 (B).](upload/image/phospholipids-analysis-service-4.jpg) The MS/ MS spectrum of PI molecular species resulting after collisional activation of the [M-H]− ions at m/z 885.5 (A) and 835.5 (B).
The MS/ MS spectrum of PI molecular species resulting after collisional activation of the [M-H]− ions at m/z 885.5 (A) and 835.5 (B).![The MS/MS spectrum of PS molecular species resulting after collisional activation of the [M-H]− ions at m/z 794.5 (A) and 772.5 (B).](upload/image/phospholipids-analysis-service-5.jpg) The MS/MS spectrum of PS molecular species resulting after collisional activation of the [M-H]− ions at m/z 794.5 (A) and 772.5 (B).
The MS/MS spectrum of PS molecular species resulting after collisional activation of the [M-H]− ions at m/z 794.5 (A) and 772.5 (B).![The MS/ MS spectrum of PC molecular species resulting after collisional activation of the [M+HCOO]− ions at m/z 802.5 (A) and 786.5 (B)](upload/image/phospholipids-analysis-service-6.jpg) The MS/ MS spectrum of PC molecular species resulting after collisional activation of the [M+HCOO]− ions at m/z 802.5 (A) and 786.5 (B)
The MS/ MS spectrum of PC molecular species resulting after collisional activation of the [M+HCOO]− ions at m/z 802.5 (A) and 786.5 (B)