Cell-based lipidomics analysis provides a unique advantage in comprehending the lipid profile within the context of a living organism. The choice of cell samples allows researchers to investigate lipid dynamics in response to various stimuli, environmental factors, or genetic modifications. Unlike isolated lipid extracts, cell lipidomics provides a holistic understanding of lipid metabolism, signaling, and regulatory pathways.
Why Choose Tissue Samples for Lipidomics Analysis?
1. Biological Relevance:
Cell-based lipidomics analysis offers a level of biological relevance that cannot be easily replicated by other sample types. Cells represent the fundamental units of life, and studying lipids within the context of a cellular environment provides insights into how lipids function in living organisms. This approach allows researchers to bridge the gap between in vitro and in vivo conditions, enhancing the translatability of findings to real physiological scenarios.
2. Dynamic Responses:
Cells are dynamic entities that respond to various stimuli, including changes in the extracellular environment, hormonal signals, or genetic modifications. Analyzing lipids within cell samples enables researchers to capture these dynamic responses over time. This temporal dimension is crucial for understanding how lipid profiles evolve in response to different conditions, shedding light on the adaptive nature of cellular lipid metabolism.
3. Functional Correlation:
Linking lipidomics with cellular functions is essential for unraveling the functional significance of different lipid species. Cell samples provide a platform to correlate lipid changes with specific cellular processes such as proliferation, differentiation, and apoptosis. This functional perspective enhances our understanding of the roles played by lipids in mediating cellular pathways, contributing to a more holistic comprehension of lipid biology.
4. Cellular Heterogeneity:
Cell populations are inherently heterogeneous, with different cell types coexisting within tissues. By choosing cell samples for lipidomics analysis, researchers can explore the diversity in lipid profiles among distinct cell types. This is particularly valuable in understanding tissue-specific lipid distributions and unraveling the complexities of lipid metabolism within multicellular organisms.
5. Reproducibility and Standardization:
Cell cultures offer a controlled and reproducible system for lipidomics analysis. Standardized cell culture techniques enable researchers to maintain consistent experimental conditions, minimizing variability introduced by external factors. This reproducibility is crucial for obtaining reliable and comparable lipidomic data, facilitating the validation of results across different studies.
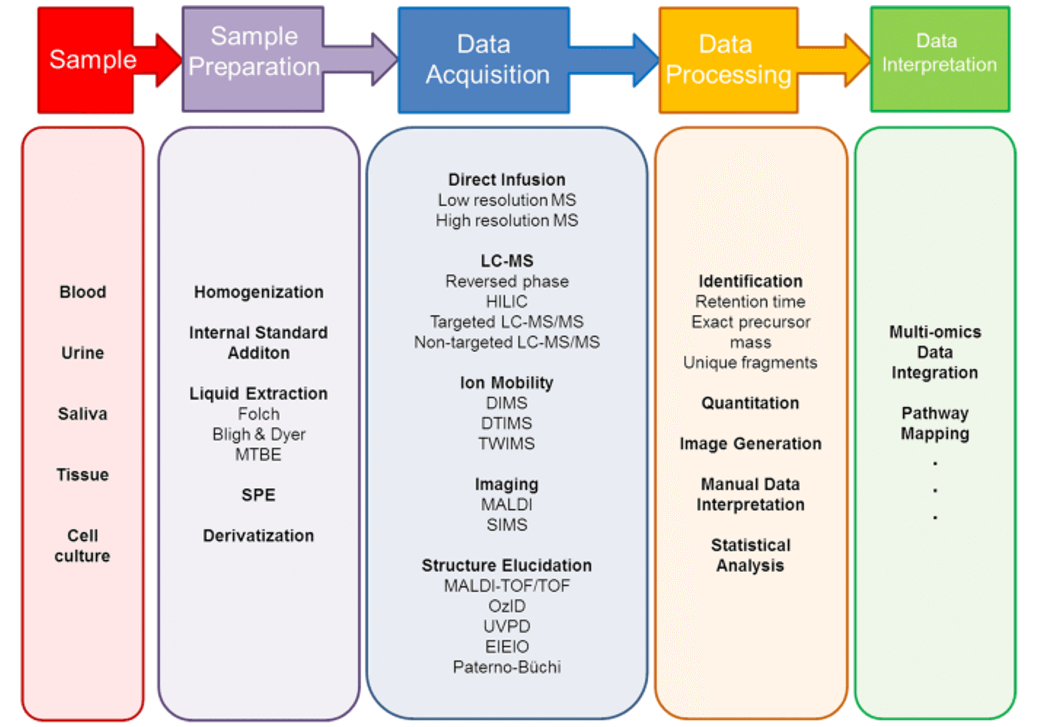
Cell Lipidomics Analysis Process
Sample Collection and Preparation:
The journey begins with the collection of cell samples. Whether adherent or suspension cells, our experts ensure proper handling and preservation to maintain cellular integrity. Efficient cell lysis techniques are employed to extract lipids effectively, setting the stage for downstream analysis.
Lipid Extraction:
Creative Proteomics employs robust lipid extraction protocols to ensure the recovery of a diverse range of lipids. Our methods are optimized to cover the lipidome comprehensively, capturing both abundant and low-abundance lipid species.
Mass Spectrometry-Based Analysis:
a. Triple Quadrupole Mass Spectrometers (QqQ):
Applied for targeted lipid analysis, QqQ mass spectrometers offer high sensitivity and specificity. This enables the precise quantification of specific lipid species, providing detailed insights into targeted pathways or lipid classes.
b. Time-of-Flight Mass Spectrometers (TOF-MS):
Utilized for untargeted lipid profiling, TOF-MS facilitates accurate mass determination. This approach allows for the identification and quantification of a broad range of lipid species without predefined targets.
c. Ion Trap Mass Spectrometers:
Ideal for the analysis of complex lipid mixtures, ion trap mass spectrometers excel in capturing detailed structural information. This is particularly valuable for deciphering the complexity of lipid isomers and closely related species.
Data Processing and Analysis:
Our data processing pipeline involves advanced bioinformatics tools. Lipid identification, quantification, and statistical analysis are performed to extract meaningful insights from the vast datasets generated during the mass spectrometry analysis.
Results Interpretation and Reporting:
The final step encompasses the interpretation of results by our expert team. A comprehensive report is generated, detailing the identified lipid species, their relative abundances, and any significant findings. This report serves as a valuable resource for researchers seeking a deeper understanding of cellular lipidomics.
 Workflow of Cell Lipidomics Service
Workflow of Cell Lipidomics Service
Advantages of Our Analytical Techniques:
- High Sensitivity: Our mass spectrometry techniques offer exceptional sensitivity, enabling the detection of low-abundance lipid species.
- Specificity: Targeted approaches using QqQ mass spectrometers ensure specific identification and quantification of desired lipid classes.
- Comprehensive Profiling: Utilizing both targeted and untargeted approaches allows us to provide a comprehensive view of the cellular lipidome.
- Structural Insight: Ion trap mass spectrometers contribute to the detailed structural characterization of complex lipid mixtures.
Sample Requirements for Cell Lipidomics
| Sample Type | Sample Collection | Cell Lysis Method | Recommended Cell Quantity | Additional Considerations |
|---|
| Tissues | Homogenization for cell release | Tissue-specific lysis protocols | 10-50 mg tissue | Tissue-specific considerations; avoid contamination during handling. |
| Cell Culture | Harvesting with trypsin or other methods | Specialized buffers for efficient lysis | Varies based on culture dish size | Consistent culture conditions and avoiding contamination are crucial. |
| Suspension Cells | Proper centrifugation and washing | Specialized detergents for lysis | 1-5 x 10^6 cells | Be aware of cell aggregation issues and optimize washing steps. |
| Adherent Cells | Gentle detachment to preserve integrity | Efficient lysis buffer application | 1-2 x 10^6 cells | Consideration of the cell culture substrate and detachment method. |
| Primary Cells | Careful isolation and purification | Enzymatic or mechanical dissociation | 0.5-1 x 10^6 cells | Attention to the isolation process to minimize cellular stress. |
Data Deliverables for Cell Lipidomics Analysis
| Data Deliverables | Description |
|---|
| Lipid Identification Report | - Detailed information on identified lipid species. |
| - Classification based on lipid categories (e.g., phospholipids, glycerolipids, sphingolipids). |
| - Precise mass and retention time of each identified lipid. |
| Lipid Quantification Data | - Quantitative data for each identified lipid species. |
| - Relative abundance or concentration levels. |
| Lipid Pathway Analysis | - Interpretation of lipid profiles in the context of cellular pathways. |
| - Correlation between lipid changes and cellular functions. |
| Statistical Analysis | - Statistical tests to assess the significance of changes in lipid abundance. |
| - Heatmaps or clustering analysis for visualizing trends. |
| Sample-to-Sample Variability | - Assessment of variability between different samples. |
| - Principal Component Analysis (PCA) plots or similar visualizations. |
| Interactive Data Visualization | - User-friendly visualizations to explore lipidomics data interactively. |
| - Graphs, charts, and plots for easy interpretation. |
| Customized Data Analysis | - Tailored analyses based on specific research questions or goals. |
| - Additional analyses as per researcher requirements. |
| Raw Data Files | - Access to raw mass spectrometry data files. |
| - Essential for verification, further analysis, or integration with other datasets. |
| Comprehensive Report | - A detailed report summarizing key findings and interpretations. |
| - Recommendations for further experiments or analyses. |
| Expert Consultation | - Availability of experts for consultation to address any questions or concerns. |
| - Assistance in interpreting complex data patterns. |
* Our services can only be used for research purposes and Not for clinical use.
Services:
Resource:
Platform:


 Workflow of Cell Lipidomics Service
Workflow of Cell Lipidomics Service