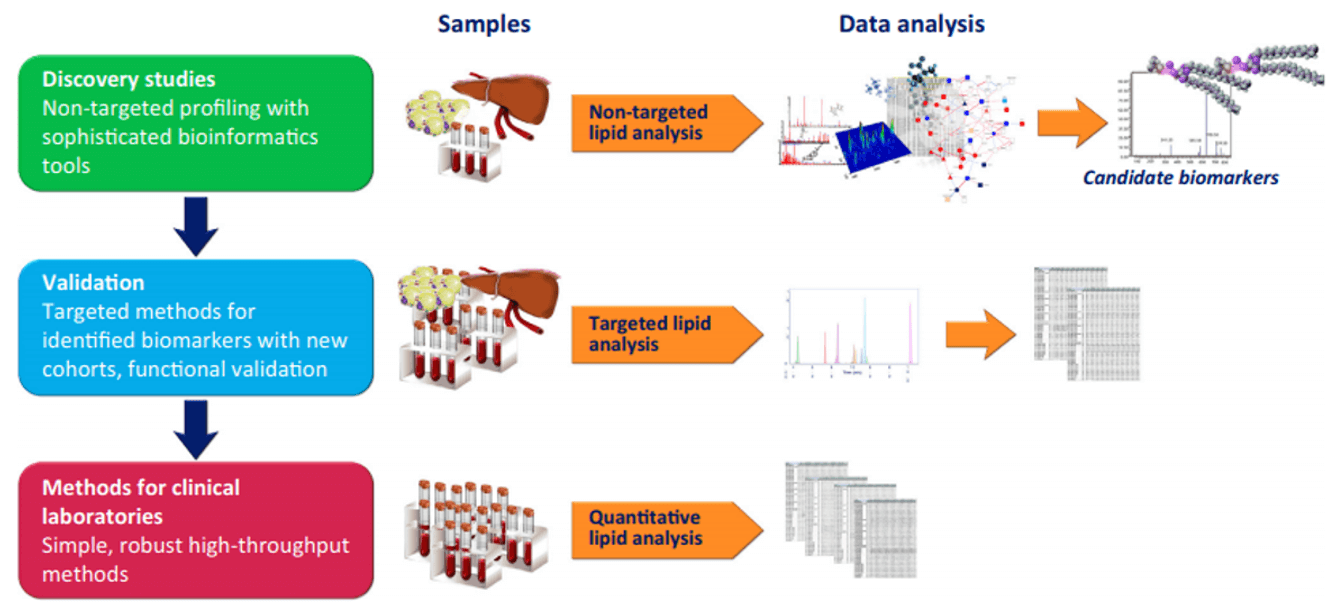
Isofurans Analysis Service
Creative Proteomics has developed a reliable and reproducible method using highly sensitive LC-MS/MS method for the rapid identification and quantification of Isofurans in different sample types, which can satisfy the needs of academic and industrial study in your lab.
Overview
Oxidative stress continues to be linked to an ever-increasing number of dysfunctional states in vivo, of which atherosclerosis, neurodegenerative diseases, cancer, inflammation, and the normal aging process are only a few examples. The deleterious effects of oxidative stress that contribute to disease most frequently manifest as chemical modification of proteins, nucleic acids, and lipids. Assessment of the role of oxidative stress in the pathogenesis or manifestations of any disease state requires a sensitive and specific in vivo marker of oxidant injury.
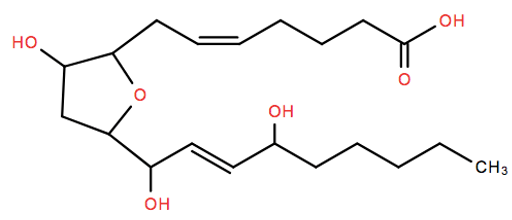
Some studies recently reported the discovery of isofurans, novel products of free radical-induced peroxidation of arachidonic acid that exhibit favored formation with increasing oxygen concentrations. Parkinson's disease is presented as a disease model involving mitochondrial dysfunction, a situation in which quantification of isofurans can provide a uniquely sensitive indicator of oxidant injury. Measurement of isofurans has also provided unexpected insights into the earliest events in hyperoxic lung injury, an important clinical problem in which measurement of isofurans might prove to be uniquely valuable in the evaluation of approaches to limit this injury.
Currently, a reliable and reproducible method using highly sensitive LC-MS/MS platform for the identification and quantification of isofurans in different sample types has been established by the scientists at Creative Proteomics. The combination of MS instrument and liquid chromatography (LC) can effectively separate target lipid molecules from complex samples.
Why Choose Us?
- We use liquid chromatography hyphenated to triple quadrupole mass spectrometers for our isofurans targeted lipidomics services, to provide high-specificity, high-selectivity, and high-reproducibility detection and quantitation of isofurans.
- Compatible with a variety of types of samples, ranging from all kinds of tissues and serum to urine.
- Creative Proteomics provides a comprehensive visual report of the results. The report will help to answer your study questions, and hopefully create a few new ones to follow.
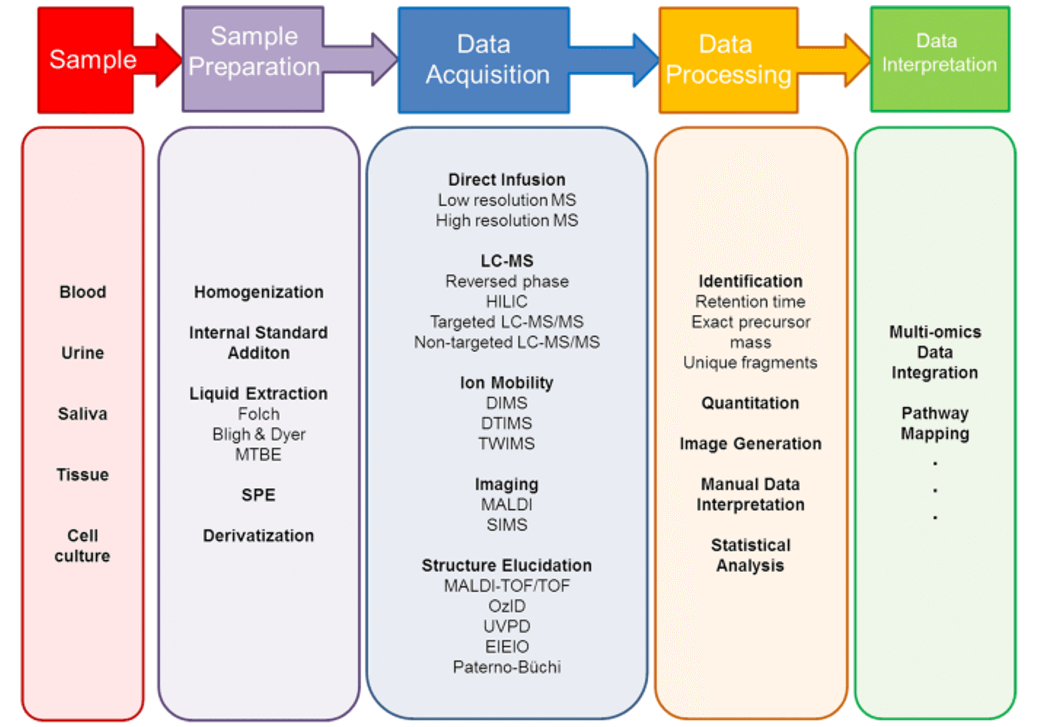
Our Isofurans Analysis Service
 Fig 2. The workflow of isofurans analysis service
Fig 2. The workflow of isofurans analysis service
Structural details: species level
Mode: MRM
Precision: pmol & mol%
Delivery time: 2-4 weeks
Sample requirements
Blood/plasma: 100-200 ul/sample
Urine: 200 ul-1 ml/sample
Tissue: 10-50 mg/ sample
- After the sample is collected, it will be stored at -80°C immediately and transported by dry ice.
- Tubes should be clearly labeled with sample identifiers and include a hard copy of the list of samples to accompany the shipment. Please include all available sample information when shipping samples.
Deliverables
- Experimental procedure
- Parameters of liquid chromatography and MS
- Purity analysis report
- MS raw data files and MS data quality checks
- Hexylceramide quantitative result data
- Custom analysis report
Based on a highly stable, reproducible and highly sensitive separation, characterization, identification and quantitative analysis system, combined with LC-MS/MS, Creative Proteomics provides reliable, fast and cost-effective Isofurans analysis services. If you have any questions about our Isofurans targeted lipidomics services, please contact us.
Reference:
- Fessel JP.; Jackson Roberts L. Isofurans: novel products of lipid peroxidation that define the occurrence of oxidant injury in settings of elevated oxygen tension. Antioxid Redox Signal. 2005 Jan-Feb;7(1-2):202-9.
* Our services can only be used for research purposes and Not for clinical use.
Services:
Resource:
Platform:



 Fig 2. The workflow of isofurans analysis service
Fig 2. The workflow of isofurans analysis service