Lipidomics in the Identification of Candidate Drugs
Identification of novel drug candidates often involves the screening of thousands of chemical compounds. In the case of lipid metabolizing enzymes, MS-based lipidomics holds great potential as it enables simultaneous and highly selective analysis of natural intermediates and end products, as well as side products due to off-target effects. However, standard lipidomic analyses have a limited throughput due to MS sample preparation involving liquid extractions, chromatographic separations, sample injection and post-MS-data processing.
Recent advances in MS instrumentation and particularly in the automation of pre- and post-MS handling of samples and data can dramatically increase the throughput of the analysis, enabling the use of MS-based lipidomics for larger compound screening projects. Particularly, the capacity of lipid extraction can be substantially increased by the use of robotic liquid handling stations. Sample injection can also be automated by using robotic sample injection and ionization devices.
One example of such systems is the TriVersa NanoMate (Advion), which is routinely used in Creative Proteomics. It employs microfluidics chips containing an array of up to 384 nanoelectrospray nozzles per chip, allowing reproducible sample injection and spraying, while avoiding cross-contamination between samples. Shotgun lipidomics is the approach of choice for high throughput applications.
Recently, high throughput mass spectrometry devices with or without parallel processing capacities have become available allowing unattended analysis of thousands of samples. Also data analysis can be automated and software can be equipped with built-in profile or pattern recognition. As each lipid metabolic enzyme possesses specific activities, unique changes in profiles are generated upon chemical inhibition or elimination of a specific enzyme. By comparing lipid profiles generated in compound screenings with available profiles of selective inhibition or knockdown of individual enzymes or their regulators, active compounds can be identified and their targets be predicted.
Case
Identification of drugs and drug targets based on the unique changes in lipid profiles each compound produces. Cancer cells (HCT116) treated with different compounds, for example an SCD1 inhibitor, an ELOVL6 inhibitor or an ACACA inhibitor show unique and characteristic changes in lipid profiles.
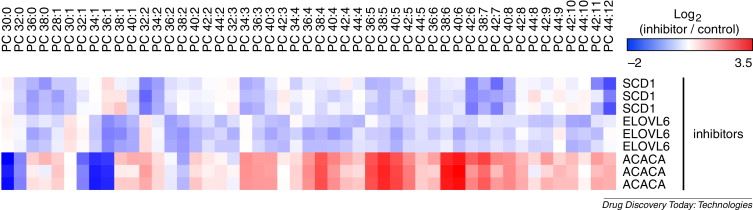 Fig1. HCT116 treated with different compounds shows unique and characteristic changes in lipid profiles (Dehairs, J.; et al. 2015)
Fig1. HCT116 treated with different compounds shows unique and characteristic changes in lipid profiles (Dehairs, J.; et al. 2015)
Creative Proteomics identifies effective drugs based on the unique changes in lipid profiles each compound produces. Particularly, the capacity of lipid extraction can be substantially increased by the use of robotic liquid handling stations. Sample injection can also be automated by using robotic sample injection and ionization devices.
Available Services
How we work in 7 easy steps

Creative Proteomics provides a perfect lipidomics platform for the identification of candidate drugs. If you have any special needs for the identification of candidate drugs, please contact us. Let us know what you need and we will accommodate you. We look forward to working with you in the future.
Reference:
- Dehairs, J.; et al. Lipidomics in drug development. Drug Discovery Today: Technologies. 2015, 13, 33-38.
* Our services can only be used for research purposes and Not for clinical use.


 Fig1. HCT116 treated with different compounds shows unique and characteristic changes in lipid profiles (Dehairs, J.; et al. 2015)
Fig1. HCT116 treated with different compounds shows unique and characteristic changes in lipid profiles (Dehairs, J.; et al. 2015)