Lipidomics in Target Discovery
With more than 180,000 different molecular species, lipids form a vast and extraordinary class of biomolecules. Each of these lipids has a unique chemical structure and biophysical properties. Functioning as structural components of membranes, as medium for energy storage, as anchor for proteins, or as intra- and inter-cellular signaling molecules, lipids play diverse roles and are involved in numerous biological processes.
The total lipid composition of an organism or specimen thereof is referred to as the lipidome. The lipidome is affected by the diet and is further controlled by a large and diverse family of lipid metabolic enzymes, which are tightly regulated by both central and cell-autonomous control mechanisms. Not surprisingly, many human diseases, including common conditions such as obesity, diabetes, cardiovascular disease, inflammatory disease, cancer and neurodegeneration involve changes in lipids and in lipid metabolic pathways. Likewise, an increasing number of drugs have been or are being developed that directly or indirectly target lipid metabolism. Established or emerging targets include enzymes involved in the synthesis and metabolism of cholesterol, fatty acids, phospholipids, sphingolipids and triglycerides. Also their regulators, including key signaling proteins and transcription factors have been identified as potential targets in the pathologies mentioned above.
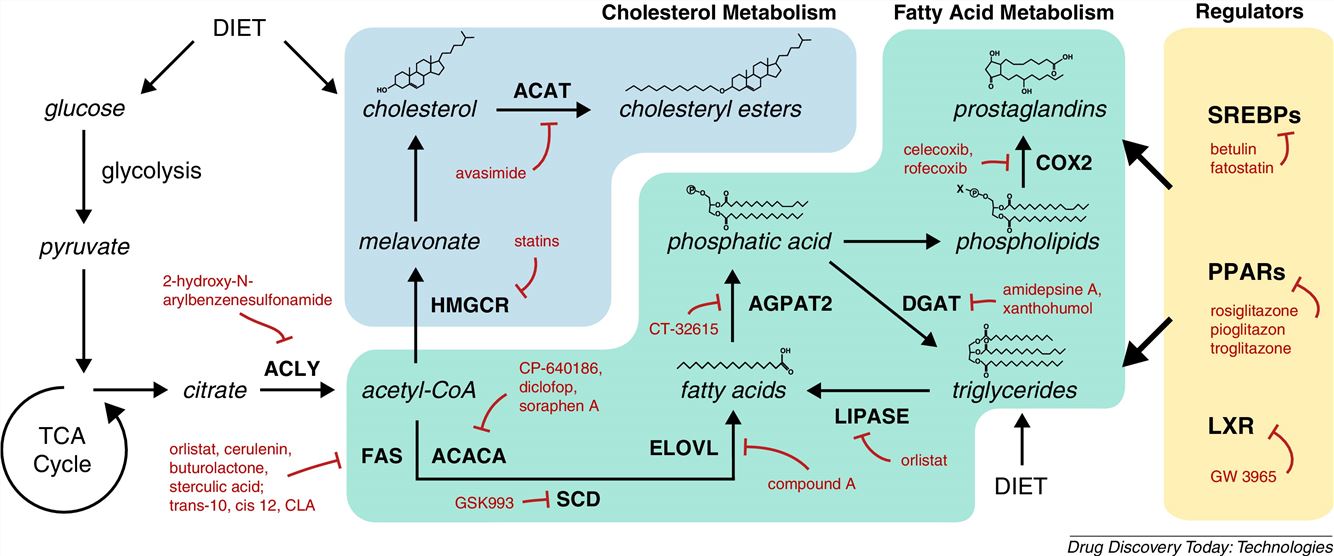 Fig1. Lipid metabolic enzymes as targets for drug development (Dehairs, J.; et al, 2015)
Fig1. Lipid metabolic enzymes as targets for drug development (Dehairs, J.; et al, 2015)
Since its development, MS-based lipidomics has been applied for the analysis of lipids in numerous disease conditions. Analysis can be carried out on tissues or body fluids of diseased and healthy human subjects as well as on disease models involving animals or cultured cells. Both LC–MS and shotgun lipidomics have been very successful in this context and have led to the identification of hitherto unknown changes in lipid profiles. Combined with transcriptome analysis and other functional genetics tools, these findings have led to new insights in disease mechanisms and to the discovery of potential new targets for drug development.
Case
Using a mass spectrometry (MS)‐based phospholipidomics approach, a study profiled 179 phospholipid species in malignant and matched non-malignant lung tissue of 162 NSCLC patients. The study identified 91 phospholipid species that were differentially expressed in cancer versus non-malignant tissues. The most prominent changes included a decrease in sphingomyelins (SMs) and an increase in specific phosphatidylinositols (PIs). The identified lipid markers that can discriminate tumor versus normal tissue and different NSCLC subtypes with an AUC (area under the ROC curve) of 0.999 and 0.885, respectively.
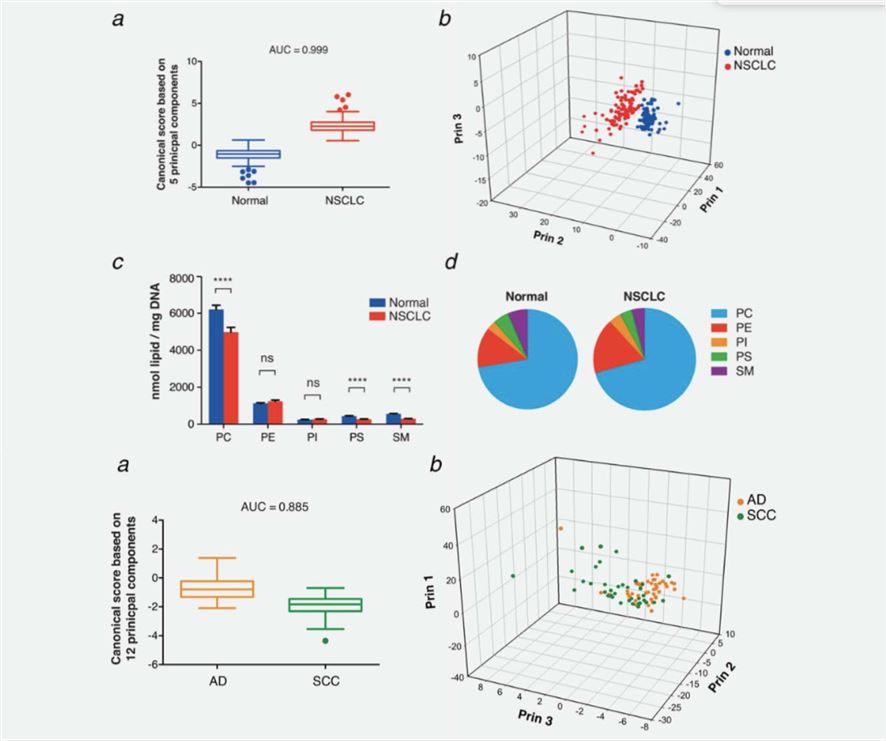 Fig2. Phospholipid profiles discriminate NSCLC versus normal tissues and AD versus SCC tumors (Marien, Eyra.; et al, 2015)
Fig2. Phospholipid profiles discriminate NSCLC versus normal tissues and AD versus SCC tumors (Marien, Eyra.; et al, 2015)
Several sphingolipids and sphingolipid hydrolases were analyzed in brain samples from AD patients and age-matched normal individuals. The study found a pattern of elevated acid sphingomyelinase (ASM) and acid ceramidase (AC) expression in AD, leading to a reduction in sphingomyelin and elevation of ceramide.
MS-based lipidomics has great potential in all these areas. Creative Proteomics provides lipidomics services for drug target discovery.
Related Featured Services
How we work in 7 easy steps

Creative Proteomics provides a perfect lipidomics platform for drug target discovery. If you have any special needs for drug target discovery, please contact us. Let us know what you need and we will accommodate you. We look forward to working with you in the future.
References:
- Dehairs, J.; et al. Lipidomics in drug development. Drug Discovery Today: Technologies. 2015, 13, 33-38.
- Marien, Eyra.; et al. Non-small cell lung cancer is characterized by dramatic changes in phospholipid profiles. International Journal of Cancer. 2015, 137.7: 1539-1548.
* Our services can only be used for research purposes and Not for clinical use.


 Fig1. Lipid metabolic enzymes as targets for drug development (Dehairs, J.; et al, 2015)
Fig1. Lipid metabolic enzymes as targets for drug development (Dehairs, J.; et al, 2015) Fig2. Phospholipid profiles discriminate NSCLC versus normal tissues and AD versus SCC tumors (Marien, Eyra.; et al, 2015)
Fig2. Phospholipid profiles discriminate NSCLC versus normal tissues and AD versus SCC tumors (Marien, Eyra.; et al, 2015)