Lipidomics in Food Origin Identification
Food origin is one of the most concerned authenticity issues among consumers all over the world and it is considered as an essential indicator of food quality. The information regarding a food product during this journey typically includes its plant or animal strains, geographical conditions, planting or rearing methods, processing and storage technologies. Consequently, robust and effective analytical methodologies are required to capture these features and MS-based lipidomics strategies play an important role in this respect.
| Varieties | The plant or animal strain of a food ingredient may be considered as the start of food traceability. By analogy to other metabolites, lipids in biomaterials consumed as food ingredients are also genotype-dependent, which means that they vary among different species, varieties or cultivars. The analysis of lipid composition in food products not only gives comprehensive information to evaluate their nutritional values, but also facilitates the differentiation of their species, varieties or cultivars, and further food fraud identification. |
| Geographical Origin | Geographical origin of a food product is also a concern of consumers. Assuring the consistence of the geographical origin of a food product and its label is a prerequisite for food fraud prevention. In this area, untargeted MS-based lipidomics has taken the leading position and has been successfully ap plied to identify the geographical origins of saffron, coix seeds, goat milk and meat. |
| Planting or Rearing Systems | It is universally believed that the farming, rearing and feeding practices can potentially affects the metabolites including lipids profiles in resulting agro-foods. In this context, several studies employing MS-based lipidomics approaches have investigated the differences between wild and farmed products. |
| Processing and storage methods | Lipids are one of the three macronutrients and also unstable components in many foodstuffs, whose changes are a major cause of quality deterioration of foods. Therefore, the exhaustive analysis of lipids and their alterations during different production steps will help to select tailored modes of processing and storage for various food matrices or optimize the production parameters. |
With decades of operational experience and technology platform, Creative Proteomics provides reliable, rapid, and cost-effective untargeted lipidomics and targeted lipidomics services based on GC/LC-MS and shot-gun methods for Food Origin Identification.
Case
Thermal processing methods have important effects on food lipids. In this work, ultra-high-performance liquid chromatography-Q-Extractive Orbitrap mass spectrometry and lipidsearch software were applied to analyze effect of three types of thermal processing methods on the lipidomics profile of tilapia fillets.
The results demonstrated three types of thermal processing methods had different effects on lipidomics profile differences of tilapia fillets. A total of eight lipid species variables (LPS, LPG, LPI, DG, LPC, TG, LPE, and Cer) and 137 individual lipids variables showed significant differences among raw, steamed, boiled, and roasted tilapia fillets.
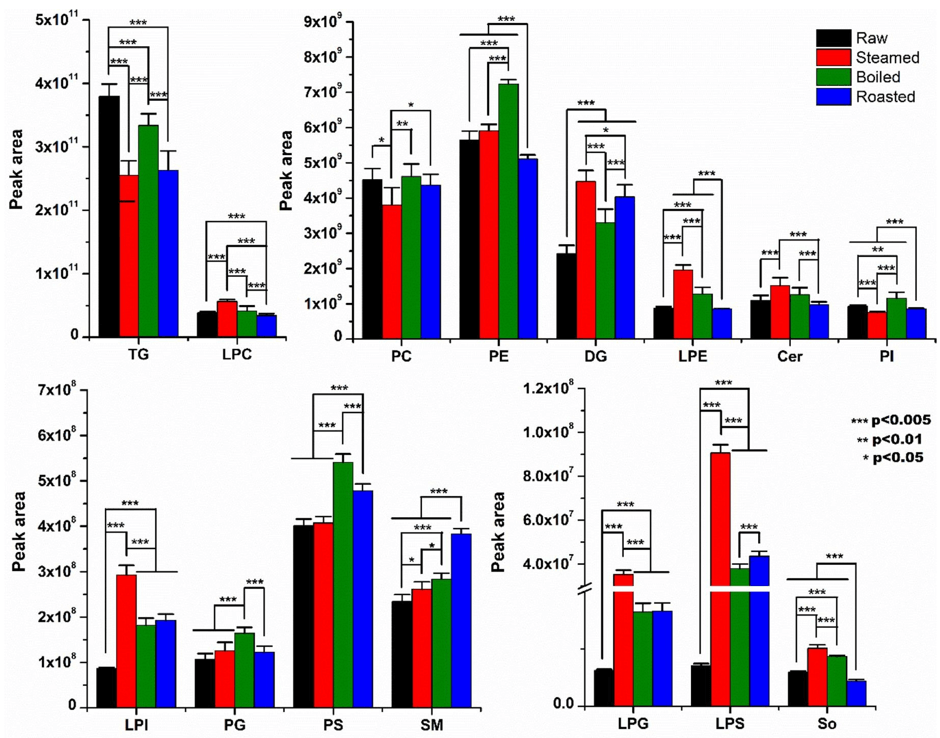 Fig1. The analyzed peak areas in raw and processed tilapia fillets (Shi, Cuiping.; et al, 2019)
Fig1. The analyzed peak areas in raw and processed tilapia fillets (Shi, Cuiping.; et al, 2019)
Related Featured Services
How we work in 7 easy steps

If you have any questions about our lipidomics services in food origin identification, please contact us. With our highly experienced scientific team, advanced techniques and equipment, we can tailor our services according to your needs.
Reference:
- Shi, Cuiping.; et al. Effect of three types of thermal processing methods on the lipidomics profile of tilapia fillets by UPLC-Q-Extractive Orbitrap mass spectrometry. Food chemistry. 2019, 298: 125029.
* Our services can only be used for research purposes and Not for clinical use.


 Fig1. The analyzed peak areas in raw and processed tilapia fillets (Shi, Cuiping.; et al, 2019)
Fig1. The analyzed peak areas in raw and processed tilapia fillets (Shi, Cuiping.; et al, 2019)