Antibiotic Resistance Research
The severity of the ongoing antibiotic resistance crisis is leading to novel and creative approaches to characterize and classify bacteria. While the field has largely been driven by genomics and transcriptomics, there is growing use of mass spectrometry–based lipidomics, towards the study of microbial systems. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF) platforms has been established for the identification of microbial species. Recent works on bacterial lipid analysis by MS have demonstrated that lipidomic signatures of microorganisms may be equally or more powerful than those based on proteomics.
On the other hand, there remains a need to develop entirely new or significantly modified antimicrobial therapeutics that are successful when other antibiotics no longer work. The development of new antimicrobial therapeutics necessitates a deep understanding of the mechanisms that enable microbial pathogens to evade treatment. Several recent works using MS-based lipidomics have demonstrated that lipid profiles of antibiotic-resistant strains differ significantly from their susceptible counterparts when the target of the antibiotic is directly associated with membrane lipids and when it is not.
Case
Staphylococcus aureus resistance to antibiotics is a significant clinical problem worldwide. In this study, an untargeted lipidomics approach was used to compare the lipid fingerprints of S. aureus clinical isolates that are resistant and sensitive to antibiotics. High-performance liquid chromatography coupled with time-of-flight mass spectrometry was employed to rapidly and comprehensively analyze bacterial lipids.
Chemometric and statistical analyses of the obtained lipid fingerprints revealed variations in several lipid groups between S. aureus strains resistant and sensitive to tested antibiotics including methicillin, gentamicin, ciprofloxacin, erythromycin, and fusidic acid. The levels of identified monoglycosyldiacylglycerol, phosphatidylglycerol, and diglycosyldiacylglycerol lipid groups were found to be upregulated in antibiotic-resistant S. aureus strains, whereas the levels of diacylglycerol lipid groups were downregulated.
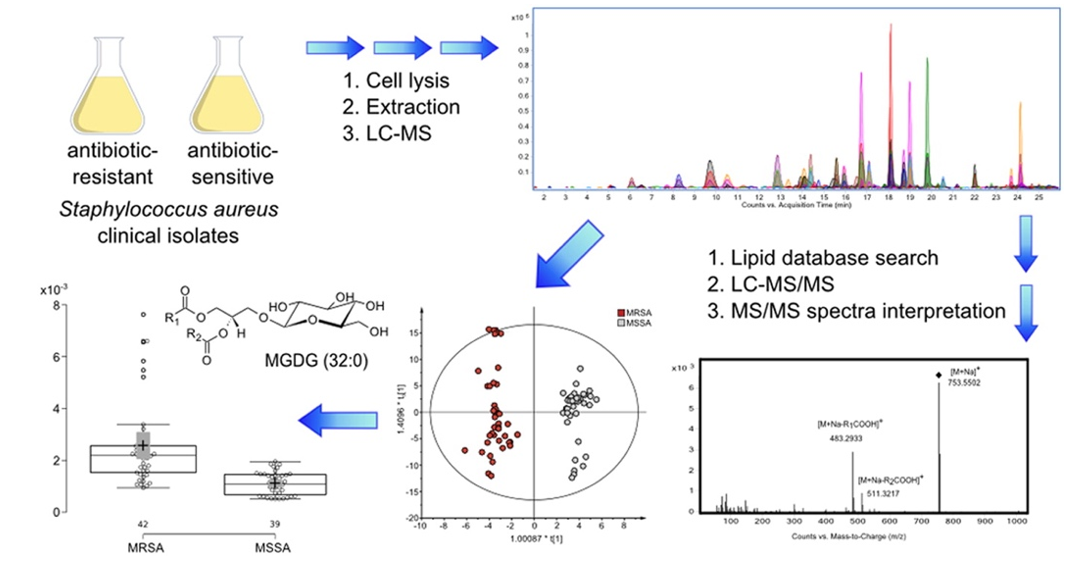 Fig1. Differences in the lipid pattern among staphylococcus aureus resistant and sensitive to Antibiotics (Hewelt-Belka, Weronika.; et al, 2016)
Fig1. Differences in the lipid pattern among staphylococcus aureus resistant and sensitive to Antibiotics (Hewelt-Belka, Weronika.; et al, 2016)
Differences in the lipid patterns between sensitive and resistant S. aureus strains suggest that antibiotic susceptibility may be associated with the lipid composition of bacterial cells. The lipids that were found to significantly differ between antibiotic-resistant and antibiotic-sensitive clinical isolates are involved in the biosynthesis of major S. aureus membrane lipids and lipoteichoic acid. This study indicates that S. aureus lipid biosynthesis pathways should be explored further to better understand the mechanism of antibiotic resistance in S. aureus strains.
The correlation between bacterial lipid composition and development of resistance is well-established. With decades of operational experience and technology platform, Creative Proteomics provides reliable, rapid, and cost-effective microorganisms lipidomics services based on MALDI-TOF methods for antibiotic resistance research.
Related Featured Services
How we work in 7 easy steps

If you have any questions about our lipidomics services for antibiotic resistance research, please contact us. With our highly experienced scientific team, advanced techniques and equipment, we can tailor our services according to your needs.
References:
- Appala, Keerthi.; et al. Recent applications of mass spectrometry in bacterial lipidomics. Analytical and Bioanalytical Chemistry. 2020, 412.24: 5935-5943.
- Hewelt-Belka, Weronika.; et al. Untargeted lipidomics reveals differences in the lipid pattern among clinical isolates of Staphylococcus aureus resistant and sensitive to antibiotics. Journal of proteome research. 2016, 15.3 (2016): 914-922.
* Our services can only be used for research purposes and Not for clinical use.
Applications:


 Fig1. Differences in the lipid pattern among staphylococcus aureus resistant and sensitive to Antibiotics (Hewelt-Belka, Weronika.; et al, 2016)
Fig1. Differences in the lipid pattern among staphylococcus aureus resistant and sensitive to Antibiotics (Hewelt-Belka, Weronika.; et al, 2016)